Reconsideration of dynamic force spectroscopy analysis of streptavidin-biotin interactions
- PMID: 20559507
- PMCID: PMC2885099
- DOI: 10.3390/ijms11052134
Reconsideration of dynamic force spectroscopy analysis of streptavidin-biotin interactions
Abstract
To understand and design molecular functions on the basis of molecular recognition processes, the microscopic probing of the energy landscapes of individual interactions in a molecular complex and their dependence on the surrounding conditions is of great importance. Dynamic force spectroscopy (DFS) is a technique that enables us to study the interaction between molecules at the single-molecule level. However, the obtained results differ among previous studies, which is considered to be caused by the differences in the measurement conditions. We have developed an atomic force microscopy technique that enables the precise analysis of molecular interactions on the basis of DFS. After verifying the performance of this technique, we carried out measurements to determine the landscapes of streptavidin-biotin interactions. The obtained results showed good agreement with theoretical predictions. Lifetimes were also well analyzed. Using a combination of cross-linkers and the atomic force microscope that we developed, site-selective measurement was carried out, and the steps involved in bonding due to microscopic interactions are discussed using the results obtained by site-selective analysis.
Keywords: atomic force microscopy; dynamic force spectroscopy; site-selective analysis; streptavidin-biotin interaction.
Figures





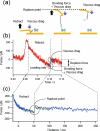


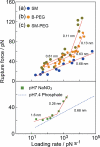



References
-
- Badjić JD, Balzani V, Credi A, Silvi S, Stoddart JF. A molecular elevator. Science. 2004;303:1845–1849. - PubMed
-
- Kay ER, Leigh DA, Zerbetto F. Synthetic molecular motors and mechanical machines. Angew. Chem.: Int. Ed. 2007;46:72–192. - PubMed
-
- Ariga K, Hill JP, Endo H. Developments in molecular recognition and sensing at interfaces. Int. J. Mol. Sci. 2007;8:864–883.
-
- Yasuda S, Okutsu Y, Suzuki I, Shinohara K, Komiyama M, Takeuchi O, Shigekawa H. Single molecular anatomy of host-guest chemistry based on atomic force microscopy study of cyclodextrin-ferrocene molecular interaction. Jpn. J. Appl. Phys. 2007;46:5614–5616.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

