Influence of the Escherichia coli oxyR gene function on lambda prophage maintenance
- PMID: 20559623
- PMCID: PMC2903704
- DOI: 10.1007/s00203-010-0596-2
Influence of the Escherichia coli oxyR gene function on lambda prophage maintenance
Abstract
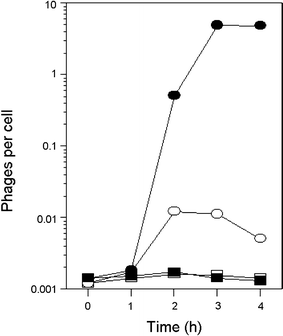
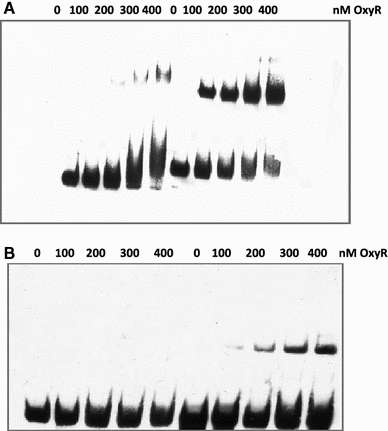
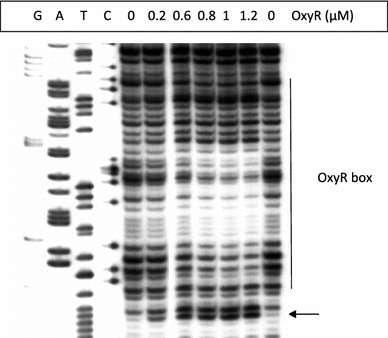
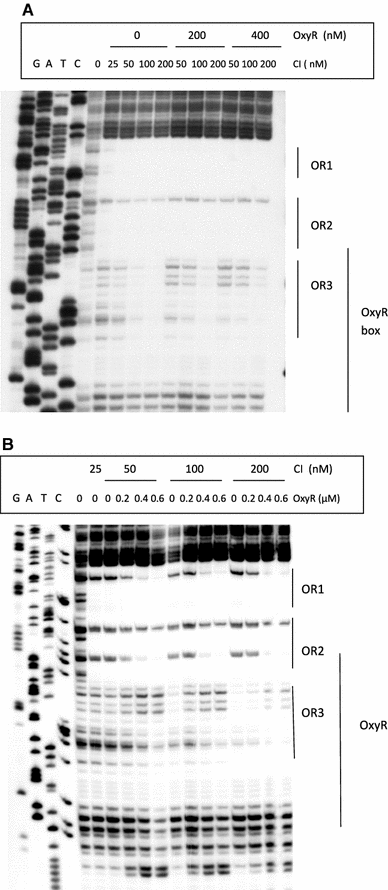
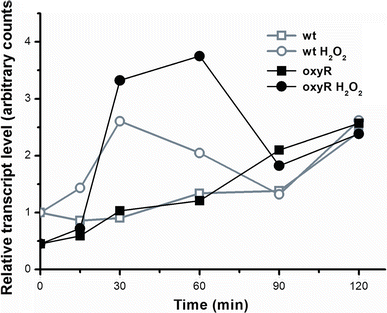
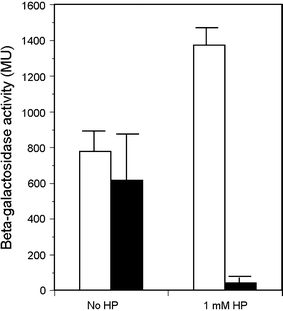
In Escherichia coli hosts, hydrogen peroxide is one of the factors that may cause induction of lambda prophage. Here, we demonstrate that H2O2-mediated lambda prophage induction is significantly enhanced in the oxyR mutant host. The mRNA levels for cI gene expression were increased in a lambda lysogen in the presence of H2O2. On the other hand, stimulation of the p(M) promoter by cI857 overproduced from a multicopy plasmid was decreased in the DeltaoxyR mutant in the presence of H2O2 but not under normal growth conditions. The purified OxyR protein did bind specifically to the p(M) promoter region. This binding impaired efficiency of interaction of the cI protein with the OR3 site, while stimulating such a binding to OR2 and OR1 sites, in the regulatory region of the p(M) promoter. We propose that changes in cI gene expression, perhaps in combination with moderately induced SOS response, may be responsible for enhanced lambda prophage induction by hydrogen peroxide in the oxyR mutant. Therefore, OxyR seems to be a factor stimulating lambda prophage maintenance under conditions of oxidative stress. This proposal is discussed in the light of efficiency of induction of lambdoid prophages bearing genes coding for Shiga toxins.
Figures

Similar articles
-
Inhibition of spontaneous induction of lambdoid prophages in Escherichia coli cultures: simple procedures with possible biotechnological applications.BMC Biotechnol. 2001;1:1. doi: 10.1186/1472-6750-1-1. Epub 2001 Apr 18. BMC Biotechnol. 2001. PMID: 11316465 Free PMC article.
-
Hydrogen peroxide-mediated induction of the Shiga toxin-converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7.FEMS Immunol Med Microbiol. 2010 Apr;58(3):322-9. doi: 10.1111/j.1574-695X.2009.00644.x. Epub 2009 Dec 10. FEMS Immunol Med Microbiol. 2010. PMID: 20070366
-
Differential efficiency of induction of various lambdoid prophages responsible for production of Shiga toxins in response to different induction agents.Microb Pathog. 2009 Dec;47(6):289-98. doi: 10.1016/j.micpath.2009.09.006. Epub 2009 Sep 15. Microb Pathog. 2009. PMID: 19761828
-
Yet another way that phage λ manipulates its Escherichia coli host: λrexB is involved in the lysogenic-lytic switch.Mol Microbiol. 2015 May;96(4):689-93. doi: 10.1111/mmi.12969. Epub 2015 Mar 16. Mol Microbiol. 2015. PMID: 25684601 Review.
-
The OxyR regulon.Antonie Van Leeuwenhoek. 1990 Oct;58(3):157-61. doi: 10.1007/BF00548927. Antonie Van Leeuwenhoek. 1990. PMID: 2256675 Review.
Cited by
-
A transcription factor from the cryptic Escherichia coli Rac prophage controls both phage and host operons.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf113. doi: 10.1093/nar/gkaf113. Nucleic Acids Res. 2025. PMID: 40037713 Free PMC article.
-
Inhibition of Shiga toxin-converting bacteriophage development by novel antioxidant compounds.J Enzyme Inhib Med Chem. 2018 Dec;33(1):639-650. doi: 10.1080/14756366.2018.1444610. J Enzyme Inhib Med Chem. 2018. PMID: 29536772 Free PMC article.
-
Gradients in gene essentiality reshape antibacterial research.FEMS Microbiol Rev. 2022 May 6;46(3):fuac005. doi: 10.1093/femsre/fuac005. FEMS Microbiol Rev. 2022. PMID: 35104846 Free PMC article. Review.
-
Inhibition of development of Shiga toxin-converting bacteriophages by either treatment with citrate or amino acid starvation.Foodborne Pathog Dis. 2012 Jan;9(1):13-9. doi: 10.1089/fpd.2011.0980. Epub 2011 Nov 2. Foodborne Pathog Dis. 2012. PMID: 22047055 Free PMC article.
-
Isothiocyanates as effective agents against enterohemorrhagic Escherichia coli: insight to the mode of action.Sci Rep. 2016 Feb 29;6:22263. doi: 10.1038/srep22263. Sci Rep. 2016. PMID: 26922906 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

