A generative model for image segmentation based on label fusion
- PMID: 20562040
- PMCID: PMC3268159
- DOI: 10.1109/TMI.2010.2050897
A generative model for image segmentation based on label fusion
Abstract
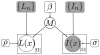
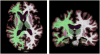
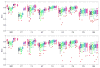
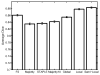
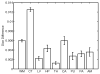
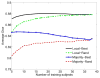
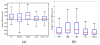
We propose a nonparametric, probabilistic model for the automatic segmentation of medical images, given a training set of images and corresponding label maps. The resulting inference algorithms rely on pairwise registrations between the test image and individual training images. The training labels are then transferred to the test image and fused to compute the final segmentation of the test subject. Such label fusion methods have been shown to yield accurate segmentation, since the use of multiple registrations captures greater inter-subject anatomical variability and improves robustness against occasional registration failures. To the best of our knowledge, this manuscript presents the first comprehensive probabilistic framework that rigorously motivates label fusion as a segmentation approach. The proposed framework allows us to compare different label fusion algorithms theoretically and practically. In particular, recent label fusion or multiatlas segmentation algorithms are interpreted as special cases of our framework. We conduct two sets of experiments to validate the proposed methods. In the first set of experiments, we use 39 brain MRI scans-with manually segmented white matter, cerebral cortex, ventricles and subcortical structures-to compare different label fusion algorithms and the widely-used FreeSurfer whole-brain segmentation tool. Our results indicate that the proposed framework yields more accurate segmentation than FreeSurfer and previous label fusion algorithms. In a second experiment, we use brain MRI scans of 282 subjects to demonstrate that the proposed segmentation tool is sufficiently sensitive to robustly detect hippocampal volume changes in a study of aging and Alzheimer's Disease.
Figures


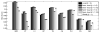
References
-
- Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726–738. - PubMed
-
- Allassonnière S, Amit Y, Trouve A. Towards a coherent statistical framework for dense deformable template estimation. J R Stat Soc B. 2007;69:3–29.
-
- Allassonnière S, Kuhn E, Trouvé A. MAP estimation of statistical deformable template via nonlinear mixed effect models: Deterministic and stochastic approaches. Math. Foundations Computat. Anat. (MFCA) Workshop MICCAI 2008 Conf; 2008.
-
- Arsigny V, Commowick O, Pennec X, Ayache N. Proc of MICCAI. Vol. 4190. New York: Springer; 2006. A log-Euclidean framework for statistics on diffeomorphisms; pp. 924–931. Lecture Notes Computer Science. - PubMed
-
- Artaechevarria X, Munoz-Barrutia A, de Solorzano CO. Efficient classifier generation and weighted voting for atlas-based segmentation: Two small steps faster and closer to the combination oracle. SPIE Med. Imag; 2008; 2008. p. 6914.
Publication types
MeSH terms
Grants and funding
- U24 RR021382/RR/NCRR NIH HHS/United States
- P41-RR14075/RR/NCRR NIH HHS/United States
- P41-RR13218/RR/NCRR NIH HHS/United States
- R01EB006758/EB/NIBIB NIH HHS/United States
- R01-NS051826/NS/NINDS NIH HHS/United States
- R01 NS052585-01/NS/NINDS NIH HHS/United States
- R01 EB001550/EB/NIBIB NIH HHS/United States
- U54 EB005149/EB/NIBIB NIH HHS/United States
- R01 NS052585/NS/NINDS NIH HHS/United States
- P41 RR013218/RR/NCRR NIH HHS/United States
- R01 RR16594-01A1/RR/NCRR NIH HHS/United States
- U24-RR021382/RR/NCRR NIH HHS/United States
- R01 RR016594/RR/NCRR NIH HHS/United States
- R01 EB006758/EB/NIBIB NIH HHS/United States
- U54-EB005149/EB/NIBIB NIH HHS/United States
- P41 RR014075/RR/NCRR NIH HHS/United States
- R01 NS051826/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

