Interactions between intracellular domains as key determinants of the quaternary structure and function of receptor heteromers
- PMID: 20562103
- PMCID: PMC2930733
- DOI: 10.1074/jbc.M110.115634
Interactions between intracellular domains as key determinants of the quaternary structure and function of receptor heteromers
Abstract
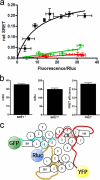
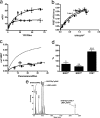
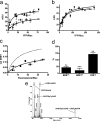
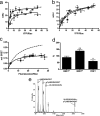
G protein-coupled receptor (GPCR) heteromers are macromolecular complexes with unique functional properties different from those of its individual protomers. Little is known about what determines the quaternary structure of GPCR heteromers resulting in their unique functional properties. In this study, using resonance energy transfer techniques in experiments with mutated receptors, we provide for the first time clear evidence for a key role of intracellular domains in the determination of the quaternary structure of GPCR heteromers between adenosine A(2A), cannabinoid CB(1), and dopamine D(2) receptors. In these interactions, arginine-rich epitopes form salt bridges with phosphorylated serine or threonine residues from CK1/2 consensus sites. Each receptor (A(2A), CB(1), and D(2)) was found to include two evolutionarily conserved intracellular domains to establish selective electrostatic interactions with intracellular domains of the other two receptors, indicating that these particular electrostatic interactions constitute a general mechanism for receptor heteromerization. Mutation experiments indicated that the interactions of the intracellular domains of the CB(1) receptor with A(2A) and D(2) receptors are fundamental for the correct formation of the quaternary structure needed for the function (MAPK signaling) of the A(2A)-CB(1)-D(2) receptor heteromers. Analysis of MAPK signaling in striatal slices of CB(1) receptor KO mice and wild-type littermates supported the existence of A(1)-CB(1)-D(2) receptor heteromer in the brain. These findings allowed us to propose the first molecular model of the quaternary structure of a receptor heteromultimer.
Figures



Similar articles
-
Moonlighting characteristics of G protein-coupled receptors: focus on receptor heteromers and relevance for neurodegeneration.IUBMB Life. 2011 Jul;63(7):463-72. doi: 10.1002/iub.473. IUBMB Life. 2011. PMID: 21698749 Review.
-
GPCR heteromers and their allosteric receptor-receptor interactions.Curr Med Chem. 2012;19(3):356-63. doi: 10.2174/092986712803414259. Curr Med Chem. 2012. PMID: 22335512 Review.
-
Interactions between calmodulin, adenosine A2A, and dopamine D2 receptors.J Biol Chem. 2009 Oct 9;284(41):28058-28068. doi: 10.1074/jbc.M109.034231. Epub 2009 Jul 24. J Biol Chem. 2009. PMID: 19632986 Free PMC article.
-
Calcium-mediated modulation of the quaternary structure and function of adenosine A2A-dopamine D2 receptor heteromers.Curr Opin Pharmacol. 2010 Feb;10(1):67-72. doi: 10.1016/j.coph.2009.10.002. Epub 2009 Nov 10. Curr Opin Pharmacol. 2010. PMID: 19896897 Free PMC article. Review.
-
Molecular integration via allosteric interactions in receptor heteromers. A working hypothesis.Curr Opin Pharmacol. 2010 Feb;10(1):14-22. doi: 10.1016/j.coph.2009.10.010. Epub 2009 Nov 26. Curr Opin Pharmacol. 2010. PMID: 19942481 Review.
Cited by
-
The Endocannabinoid System as a Target in Cancer Diseases: Are We There Yet?Front Pharmacol. 2019 Apr 5;10:339. doi: 10.3389/fphar.2019.00339. eCollection 2019. Front Pharmacol. 2019. PMID: 31024307 Free PMC article. Review.
-
Heteroreceptor Complexes Formed by Dopamine D1, Histamine H3, and N-Methyl-D-Aspartate Glutamate Receptors as Targets to Prevent Neuronal Death in Alzheimer's Disease.Mol Neurobiol. 2017 Aug;54(6):4537-4550. doi: 10.1007/s12035-016-9995-y. Epub 2016 Jul 1. Mol Neurobiol. 2017. PMID: 27370794
-
Stronger Dopamine D1 Receptor-Mediated Neurotransmission in Dyskinesia.Mol Neurobiol. 2015 Dec;52(3):1408-1420. doi: 10.1007/s12035-014-8936-x. Epub 2014 Oct 26. Mol Neurobiol. 2015. PMID: 25344317
-
Understanding the added value of g-protein-coupled receptor heteromers.Scientifica (Cairo). 2014;2014:362937. doi: 10.1155/2014/362937. Epub 2014 Apr 22. Scientifica (Cairo). 2014. PMID: 24864225 Free PMC article. Review.
-
How adenylate cyclase choreographs the pas de deux of the receptors heteromerization dance.Neuroscience. 2013 May 15;238:335-44. doi: 10.1016/j.neuroscience.2013.02.006. Epub 2013 Feb 19. Neuroscience. 2013. PMID: 23434492 Free PMC article.
References
-
- Ferré S., Ciruela F., Woods A. S., Lluis C., Franco R. (2007) Trends Neurosci. 30, 440–446 - PubMed
-
- Pin J. P., Neubig R., Bouvier M., Devi L., Filizola M., Javitch J. A., Lohse M. J., Milligan G., Palczewski K., Parmentier M., Spedding M. (2007) Pharmacol. Rev. 59, 5–13 - PubMed
-
- Dalrymple M. B., Pfleger K. D., Eidne K. A. (2008) Pharmacol. Ther. 118, 359–371 - PubMed
-
- Carriba P., Navarro G., Ciruela F., Ferré S., Casadó V., Agnati L., Cortés A., Mallol J., Fuxe K., Canela E. I., Lluís C., Franco R. (2008) Nat. Methods 5, 727–733 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

