Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings
- PMID: 20562230
- PMCID: PMC2923913
- DOI: 10.1104/pp.110.157461
Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings
Abstract
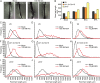
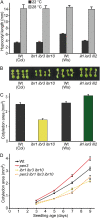
Genetic evidence in Arabidopsis (Arabidopsis thaliana) suggests that the auxin precursor indole-3-butyric acid (IBA) is converted into active indole-3-acetic acid (IAA) by peroxisomal beta-oxidation; however, direct evidence that Arabidopsis converts IBA to IAA is lacking, and the role of IBA-derived IAA is not well understood. In this work, we directly demonstrated that Arabidopsis seedlings convert IBA to IAA. Moreover, we found that several IBA-resistant, IAA-sensitive mutants were deficient in IBA-to-IAA conversion, including the indole-3-butyric acid response1 (ibr1) ibr3 ibr10 triple mutant, which is defective in three enzymes likely to be directly involved in peroxisomal IBA beta-oxidation. In addition to IBA-to-IAA conversion defects, the ibr1 ibr3 ibr10 triple mutant displayed shorter root hairs and smaller cotyledons than wild type; these cell expansion defects are suggestive of low IAA levels in certain tissues. Consistent with this possibility, we could rescue the ibr1 ibr3 ibr10 short-root-hair phenotype with exogenous auxin. A triple mutant defective in hydrolysis of IAA-amino acid conjugates, a second class of IAA precursor, displayed reduced hypocotyl elongation but normal cotyledon size and only slightly reduced root hair lengths. Our data suggest that IBA beta-oxidation and IAA-amino acid conjugate hydrolysis provide auxin for partially distinct developmental processes and that IBA-derived IAA plays a major role in driving root hair and cotyledon cell expansion during seedling development.
Figures




Similar articles
-
HOMEOBOX PROTEIN 24 mediates the conversion of indole-3-butyric acid to indole-3-acetic acid to promote root hair elongation.New Phytol. 2021 Dec;232(5):2057-2070. doi: 10.1111/nph.17719. Epub 2021 Sep 25. New Phytol. 2021. PMID: 34480752
-
Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes.Genetics. 2008 Sep;180(1):237-51. doi: 10.1534/genetics.108.090399. Epub 2008 Aug 24. Genetics. 2008. PMID: 18725356 Free PMC article.
-
Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin.Plant Cell. 2011 Mar;23(3):984-99. doi: 10.1105/tpc.111.083071. Epub 2011 Mar 15. Plant Cell. 2011. PMID: 21406624 Free PMC article.
-
Peroxisomes as a source of auxin signaling molecules.Subcell Biochem. 2013;69:257-81. doi: 10.1007/978-94-007-6889-5_14. Subcell Biochem. 2013. PMID: 23821153 Review.
-
Roles for IBA-derived auxin in plant development.J Exp Bot. 2018 Jan 4;69(2):169-177. doi: 10.1093/jxb/erx298. J Exp Bot. 2018. PMID: 28992091 Free PMC article. Review.
Cited by
-
pex5 Mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis.Plant Physiol. 2010 Dec;154(4):1602-15. doi: 10.1104/pp.110.162479. Epub 2010 Oct 25. Plant Physiol. 2010. PMID: 20974890 Free PMC article.
-
Functional Characterization of GhACX3 Gene Reveals Its Significant Role in Enhancing Drought and Salt Stress Tolerance in Cotton.Front Plant Sci. 2021 Aug 10;12:658755. doi: 10.3389/fpls.2021.658755. eCollection 2021. Front Plant Sci. 2021. PMID: 34447398 Free PMC article.
-
Factors governing cellular reprogramming competence in Arabidopsis adventitious root formation.Dev Cell. 2024 Oct 21;59(20):2745-2758.e3. doi: 10.1016/j.devcel.2024.06.019. Epub 2024 Jul 22. Dev Cell. 2024. PMID: 39043189
-
Genetic Interactions between PEROXIN12 and Other Peroxisome-Associated Ubiquitination Components.Plant Physiol. 2016 Nov;172(3):1643-1656. doi: 10.1104/pp.16.01211. Epub 2016 Sep 20. Plant Physiol. 2016. PMID: 27650450 Free PMC article.
-
PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 Regulates Lateral Root Formation via Auxin Signaling in Arabidopsis.Plant Physiol. 2014 Jun;165(2):880-894. doi: 10.1104/pp.114.239806. Epub 2014 Apr 30. Plant Physiol. 2014. PMID: 24784134 Free PMC article.
References
-
- Adham AR, Zolman BK, Millius A, Bartel B. (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in b-oxidation. Plant J 41: 859–874 - PubMed
-
- Barkawi LS, Tam YY, Tillman JA, Pederson B, Calio J, Al-Amier H, Emerick M, Normanly J, Cohen JD. (2008) A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal Biochem 372: 177–188 - PubMed
-
- Bartel B, Fink GR. (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748 - PubMed
-
- Bartel B, LeClere S, Magidin M, Zolman BK. (2001) Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid b-oxidation. J Plant Growth Regul 20: 198–216
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

