Metatranscriptomic analysis of the response of river biofilms to pharmaceutical products, using anonymous DNA microarrays
- PMID: 20562274
- PMCID: PMC2918961
- DOI: 10.1128/AEM.00873-10
Metatranscriptomic analysis of the response of river biofilms to pharmaceutical products, using anonymous DNA microarrays
Abstract
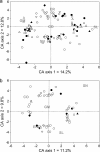
Pharmaceutical products are released at low concentrations into aquatic environments following domestic wastewater treatment. Such low concentrations have been shown to induce transcriptional responses in microorganisms, which could have consequences on aquatic ecosystem dynamics. In order to test if these transcriptional responses could also be observed in complex river microbial communities, biofilm reactors were inoculated with water from two rivers of differing trophic statuses and subsequently treated with environmentally relevant doses (ng/liter to microg/liter range) of four pharmaceuticals (erythromycin [ER], gemfibrozil [GM], sulfamethazine [SN], and sulfamethoxazole [SL]). To monitor functional gene expression, we constructed a 9,600-feature anonymous DNA microarray platform onto which cDNA from the biofilms was hybridized. Pharmaceutical treatments induced both positive and negative transcriptional responses from biofilm microorganisms. For instance, ER induced the transcription of several stress, transcription, and replication genes, while GM, a lipid regulator, induced transcriptional responses from several genes involved in lipid metabolism. SN caused shifts in genes involved in energy production and conversion, and SL induced responses from a range of cell membrane and outer envelope genes, which in turn could affect biofilm formation. The results presented here demonstrate for the first time that low concentrations of small molecules can induce transcriptional changes in a complex microbial community. The relevance of these results also demonstrates the usefulness of anonymous DNA microarrays for large-scale metatranscriptomic studies of communities from differing aquatic ecosystems.
Figures

References
-
- Beller, H. R., S. R. Kane, T. C. Legler, and P. J. J. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. - PubMed
-
- Boehm, A., S. Steiner, F. Zaehringer, A. Casanova, F. Hamburger, D. Ritz, W. Keck, M. Ackermann, T. Schirmer, and U. Jenal. 2009. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 72:1500-1516. - PubMed
-
- Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 14:45-54. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

