FADS genetic variants and omega-6 polyunsaturated fatty acid metabolism in a homogeneous island population
- PMID: 20562440
- PMCID: PMC2918459
- DOI: 10.1194/jlr.M008359
FADS genetic variants and omega-6 polyunsaturated fatty acid metabolism in a homogeneous island population
Abstract
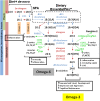
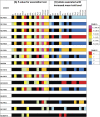
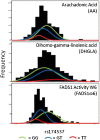
Long-chain polyunsaturated fatty acids (PUFA) orchestrate immunity and inflammation through their capacity to be converted to potent inflammatory mediators. We assessed associations of FADS gene cluster polymorphisms and fasting serum PUFA concentrations in a fully ascertained, geographically isolated founder population of European descent. Concentrations of 22 PUFAs were determined by gas chromatography, of which ten fatty acids and five ratios defining FADS1 and FADS2 activity were tested for genetic association against 16 single nucleotide polymorphisms (SNP) in 224 individuals. A cluster of SNPs in tight linkage disequilibrium in the FADS1 gene (rs174537, rs174545, rs174546, rs174553, rs174556, rs174561, rs174568, and rs99780) were strongly associated with arachidonic acid (AA) (P = 5.8 x 10(-7) - 1.7 x 10(-8)) among other PUFAs, but the strongest associations were with the ratio measuring FADS1 activity in the omega-6 series (P = 2.11 x 10(-13) - 1.8 x 10(-20)). The minor allele across all SNPs was consistently associated with decreased omega-6 PUFAs, with the exception of dihomo-gamma-linoleic acid (DHGLA), where the minor allele was consistently associated with increased levels. Our findings in a geographically isolated population with a homogenous dietary environment suggest that variants in the Delta-5 desaturase enzymatic step likely regulate the efficiency of conversion of medium-chain PUFAs to potentially inflammatory PUFAs, such as AA.
Figures

References
-
- Cordain L., Eaton S. B., Sebastian A., Mann N., Lindeberg S., Watkins B. A., O'Keefe J. H., Brand-Miller J. 2005. Origins and evolution of the Western diet: health implications for the 21st century. Am. J. Clin. Nutr. 81: 341–354. - PubMed
-
- Robson A. A. 2009. Preventing diet induced disease: bioavailable nutrient-rich, low-energy-dense diets. Nutr. Health. 20: 135–166. - PubMed
-
- Calder P. C., Albers R., Antoine J. M., Blum S., Bourdet-Sicard R., Ferns G. A., Folkerts G., Friedmann P. S., Frost G. S., Guarner F., et al. 2009. Inflammatory disease processes and interactions with nutrition. Br. J. Nutr. 101(Suppl 1): S1–S45. - PubMed
-
- Schmidt M. I., Duncan B. B. 2003. Diabesity: an inflammatory metabolic condition. Clin. Chem. Lab. Med. 41: 1120–1130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

