Savant: genome browser for high-throughput sequencing data
- PMID: 20562449
- PMCID: PMC3271355
- DOI: 10.1093/bioinformatics/btq332
Savant: genome browser for high-throughput sequencing data
Abstract
Motivation: The advent of high-throughput sequencing (HTS) technologies has made it affordable to sequence many individuals' genomes. Simultaneously the computational analysis of the large volumes of data generated by the new sequencing machines remains a challenge. While a plethora of tools are available to map the resulting reads to a reference genome, and to conduct primary analysis of the mappings, it is often necessary to visually examine the results and underlying data to confirm predictions and understand the functional effects, especially in the context of other datasets.
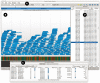
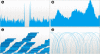
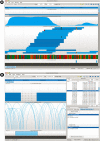
Results: We introduce Savant, the Sequence Annotation, Visualization and ANalysis Tool, a desktop visualization and analysis browser for genomic data. Savant was developed for visualizing and analyzing HTS data, with special care taken to enable dynamic visualization in the presence of gigabases of genomic reads and references the size of the human genome. Savant supports the visualization of genome-based sequence, point, interval and continuous datasets, and multiple visualization modes that enable easy identification of genomic variants (including single nucleotide polymorphisms, structural and copy number variants), and functional genomic information (e.g. peaks in ChIP-seq data) in the context of genomic annotations.
Availability: Savant is freely available at http://compbio.cs.toronto.edu/savant.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

