Network organization of the human autophagy system
- PMID: 20562859
- PMCID: PMC2901998
- DOI: 10.1038/nature09204
Network organization of the human autophagy system
Abstract
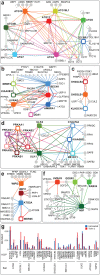
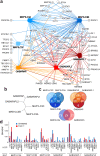
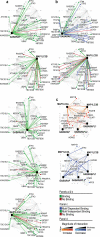
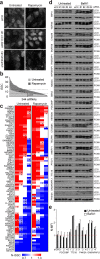
Autophagy, the process by which proteins and organelles are sequestered in autophagosomal vesicles and delivered to the lysosome/vacuole for degradation, provides a primary route for turnover of stable and defective cellular proteins. Defects in this system are linked with numerous human diseases. Although conserved protein kinase, lipid kinase and ubiquitin-like protein conjugation subnetworks controlling autophagosome formation and cargo recruitment have been defined, our understanding of the global organization of this system is limited. Here we report a proteomic analysis of the autophagy interaction network in human cells under conditions of ongoing (basal) autophagy, revealing a network of 751 interactions among 409 candidate interacting proteins with extensive connectivity among subnetworks. Many new autophagy interaction network components have roles in vesicle trafficking, protein or lipid phosphorylation and protein ubiquitination, and affect autophagosome number or flux when depleted by RNA interference. The six ATG8 orthologues in humans (MAP1LC3/GABARAP proteins) interact with a cohort of 67 proteins, with extensive binding partner overlap between family members, and frequent involvement of a conserved surface on ATG8 proteins known to interact with LC3-interacting regions in partner proteins. These studies provide a global view of the mammalian autophagy interaction landscape and a resource for mechanistic analysis of this critical protein homeostasis pathway.
Figures


Comment in
-
Autophagy: Snapshot of the network.Nature. 2010 Jul 1;466(7302):38-40. doi: 10.1038/466038a. Nature. 2010. PMID: 20596005 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

