GLI3-dependent repression of DR4 mediates hedgehog antagonism of TRAIL-induced apoptosis
- PMID: 20562908
- PMCID: PMC2928864
- DOI: 10.1038/onc.2010.235
GLI3-dependent repression of DR4 mediates hedgehog antagonism of TRAIL-induced apoptosis
Abstract
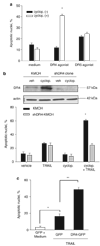
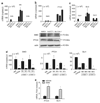
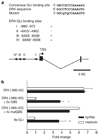
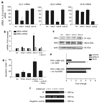
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis through its cognate receptors death receptor 4 (DR4) and death receptor 5 (DR5), preferentially in malignant cells. However, many malignant cells remain resistant to TRAIL cytotoxicity by poorly characterized mechanisms. Here, using cholangiocarcinoma cells, as a model for TRAIL resistance, we identified a role for the oncogenic Hedgehog (Hh)-GLI pathway in the regulation of TRAIL cytotoxicity. Blockade of Hh using pharmacological and genetic tools sensitizes the cells to TRAIL cytotoxicity. Restoration of apoptosis sensitivity coincided with upregulation of DR4 expression, while expression of other death effector proteins remained unaltered. Knockdown of DR4 mimics Hh-mediated resistance to TRAIL cytotoxicity. Hh regulates the expression of DR4 by modulating the activity of its promoter. Luciferase, chromatin immunoprecipitation and expression assays show that the transcription factor GLI3 binds to the DR4 promoter and Hh requires an intact GLI3-repression activity to silence DR4 expression. Finally, small interfering RNA (siRNA)-targeted knockdown of GLI3, but not GLI1 or GLI2, restores DR4 expression and TRAIL sensitivity, indicating that the Hh effect is exclusively mediated by this transcription factor. In conclusion, these data provide evidence of a regulatory mechanism, which modulates TRAIL signaling in cancer cells and suggest new therapeutic approaches for TRAIL-resistant neoplasms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. - PubMed
-
- Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. - PubMed
-
- Bigelow RL, Chari NS, Unden AB, Spurgers KB, Lee S, Roop DR, et al. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J Biol Chem. 2004;279:1197–1205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

