MassSieve: panning MS/MS peptide data for proteins
- PMID: 20564260
- PMCID: PMC2997567
- DOI: 10.1002/pmic.200900370
MassSieve: panning MS/MS peptide data for proteins
Abstract
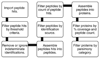
We present MassSieve, a Java-based platform for visualization and parsimony analysis of single and comparative LC-MS/MS database search engine results. The success of mass spectrometric peptide sequence assignment algorithms has led to the need for a tool to merge and evaluate the increasing data set sizes that result from LC-MS/MS-based shotgun proteomic experiments. MassSieve supports reports from multiple search engines with differing search characteristics, which can increase peptide sequence coverage and/or identify conflicting or ambiguous spectral assignments.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures

References
-
- Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics. 2005;4:1419–1440. - PubMed
-
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. - PubMed
-
- Yang X, Dondeti V, Dezube R, Maynard DM, et al. DBParser: web-based software for shotgun proteomic data analyses. J. Proteome Res. 2004;3:1002–1008. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

