Evidence for separation of HCV subtype 1a into two distinct clades
- PMID: 20565573
- PMCID: PMC2964416
- DOI: 10.1111/j.1365-2893.2010.01342.x
Evidence for separation of HCV subtype 1a into two distinct clades
Abstract
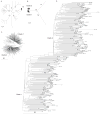
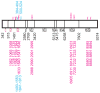
The nucleotide sequence diversity present among hepatitis C virus (HCV) isolates allows rapid adjustment to exterior forces including host immunity and drug therapy. This viral response reflects a combination of a high rate of replication together with an error-prone RNA-dependent RNA polymerase, providing for the selection and proliferation of the viruses with the highest fitness. We examined HCV subtype 1a whole-genome sequences to identify positions contributing to genotypic and phenotypic diversity. Phylogenetic tree reconstructions showed two distinct clades existing within the 1a subtype with each clade having a star-like tree topology and lacking definite correlation between time or place of isolation and phylogeny. Identification of significant phylogenetically informative sites at the nucleotide level revealed positions not only contributing to clade differentiation, but which are located at or proximal to codons associated with resistance to protease inhibitors (NS3 Q41) or polymerase inhibitors (NS5B S368). Synonymous/nonsynonymous substitution mutation analyses revealed that the majority of nucleotide mutations yielded synonymous amino acids, indicating the presence of purifying selection pressure across the polyprotein with pockets of positive selection also being detected. Despite evidence for divergence at several loci, certain 1a characteristics were preserved including the length of the alternative reading frame/F protein (ARF/F) gene, and a subtype 1a-specific phosphorylation site in NS5A (S349). Our analysis suggests that there may be strain-specific differences in the development of antiviral resistance to viruses infecting patients who are dependent on the genetic variation separating these two clades.
© 2010 Blackwell Publishing Ltd.
Conflict of interest statement
The authors have no conflict(s) of interest.
(i) This study was funded [
Figures

Similar articles
-
The presence of resistance mutations to protease and polymerase inhibitors in Hepatitis C virus sequences from the Los Alamos databank.J Viral Hepat. 2013 Jun;20(6):414-21. doi: 10.1111/jvh.12051. Epub 2013 Feb 6. J Viral Hepat. 2013. PMID: 23647958
-
HCV genotypes are differently prone to the development of resistance to linear and macrocyclic protease inhibitors.PLoS One. 2012;7(7):e39652. doi: 10.1371/journal.pone.0039652. Epub 2012 Jul 6. PLoS One. 2012. PMID: 22792183 Free PMC article.
-
[Determination of drug resistance mutations of NS3 inhibitors in chronic hepatitis C patients infected with genotype 1].Mikrobiyol Bul. 2017 Apr;51(2):145-155. doi: 10.5578/mb.53824. Mikrobiyol Bul. 2017. PMID: 28566078 Turkish.
-
NS3 protease polymorphisms and genetic barrier to drug resistance of distinct hepatitis C virus genotypes from worldwide treatment-naïve subjects.J Viral Hepat. 2016 Nov;23(11):840-849. doi: 10.1111/jvh.12503. Epub 2016 Jan 18. J Viral Hepat. 2016. PMID: 26775769 Review.
-
Hepatitis C virus resistance to protease inhibitors.J Hepatol. 2011 Jul;55(1):192-206. doi: 10.1016/j.jhep.2011.01.011. Epub 2011 Feb 1. J Hepatol. 2011. PMID: 21284949 Review.
Cited by
-
Elucidating the multi-target pharmacological mechanism of Xiaoyandina for the treatment of hepatitis C virus based on bioinformatics and cyberpharmacology studies.Medicine (Baltimore). 2025 Mar 14;104(11):e41793. doi: 10.1097/MD.0000000000041793. Medicine (Baltimore). 2025. PMID: 40101098 Free PMC article.
-
Two Distinct Hepatitis C Virus Genotype 1a Clades Have Different Geographical Distribution and Association With Natural Resistance to NS3 Protease Inhibitors.Open Forum Infect Dis. 2015 Mar 31;2(2):ofv043. doi: 10.1093/ofid/ofv043. eCollection 2015 Apr. Open Forum Infect Dis. 2015. PMID: 26213689 Free PMC article.
-
NS3 genomic sequencing and phylogenetic analysis as alternative to a commercially available assay to reliably determine hepatitis C virus subtypes 1a and 1b.PLoS One. 2017 Jul 28;12(7):e0182193. doi: 10.1371/journal.pone.0182193. eCollection 2017. PLoS One. 2017. PMID: 28753662 Free PMC article.
-
Resistance analysis of baseline and treatment-emergent variants in hepatitis C virus genotype 1 in the AVIATOR study with paritaprevir-ritonavir, ombitasvir, and dasabuvir.Antimicrob Agents Chemother. 2015 Sep;59(9):5445-54. doi: 10.1128/AAC.00998-15. Epub 2015 Jun 22. Antimicrob Agents Chemother. 2015. PMID: 26100711 Free PMC article.
-
Hepatitis C virus genetic diversity by geographic region within genotype 1-6 subtypes among patients treated with glecaprevir and pibrentasvir.PLoS One. 2018 Oct 4;13(10):e0205186. doi: 10.1371/journal.pone.0205186. eCollection 2018. PLoS One. 2018. PMID: 30286205 Free PMC article.
References
-
- Lemon SM, Walker C, Alter MJ, Yi M. Hepatitis C virus. Philadelphia, PA: Lippincott Williams & Wilkins; 2007.
-
- Vassilaki N, Mavromara P. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. The Journal of biological chemistry. 2003;278(42):40503–13. - PubMed
-
- Dhar D, Mapa K, Pudi R, Srinivasan P, Bodhinathan K, Das S. Human ribosomal protein L18a interacts with hepatitis C virus internal ribosome entry site. Archives of virology. 2006;151(3):509–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

