Comparative analysis of cis-regulation following stroke and seizures in subspaces of conserved eigensystems
- PMID: 20565733
- PMCID: PMC2902439
- DOI: 10.1186/1752-0509-4-86
Comparative analysis of cis-regulation following stroke and seizures in subspaces of conserved eigensystems
Abstract
Background: It is often desirable to separate effects of different regulators on gene expression, or to identify effects of the same regulator across several systems. Here, we focus on the rat brain following stroke or seizures, and demonstrate how the two tasks can be approached simultaneously.
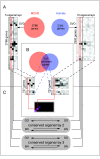
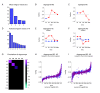
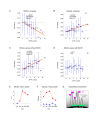
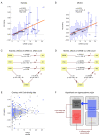
Results: We applied SVD to time-series gene expression datasets from the rat experimental models of stroke and seizures. We demonstrate conservation of two eigensystems, reflecting inflammation and/or apoptosis (eigensystem 2) and neuronal synaptic activity (eigensystem 3), between the stroke and seizures. We analyzed cis-regulation of gene expression in the subspaces of the conserved eigensystems. Bayesian networks analysis was performed separately for either experimental model, with cross-system validation of the highest-ranking features. In this way, we correctly re-discovered the role of AP1 in the regulation of apoptosis, and the involvement of Creb and Egr in the regulation of synaptic activity-related genes. We identified a novel antagonistic effect of the motif recognized by the nuclear matrix attachment region-binding protein Satb1 on AP1-driven transcriptional activation, suggesting a link between chromatin loop structure and gene activation by AP1. The effects of motifs binding Satb1 and Creb on gene expression in brain conform to the assumption of the linear response model of gene regulation. Our data also suggest that numerous enhancers of neuronal-specific genes are important for their responsiveness to the synaptic activity.
Conclusion: Eigensystems conserved between stroke and seizures separate effects of inflammation/apoptosis and neuronal synaptic activity, exerted by different transcription factors, on gene expression in rat brain.
Figures



Similar articles
-
Bayesian variable selection for gene expression modeling with regulatory motif binding sites in neuroinflammatory events.Neuroinformatics. 2006 Winter;4(1):95-117. doi: 10.1385/NI:4:1:95. Neuroinformatics. 2006. PMID: 16595861
-
Investigating the effect of dependence between conditions with Bayesian Linear Mixed Models for motif activity analysis.PLoS One. 2020 May 1;15(5):e0231824. doi: 10.1371/journal.pone.0231824. eCollection 2020. PLoS One. 2020. PMID: 32357166 Free PMC article.
-
SATB1 Mediates Long-Range Chromatin Interactions: A Dual Regulator of Anti-Apoptotic BCL2 and Pro-Apoptotic NOXA Genes.PLoS One. 2015 Sep 30;10(9):e0139170. doi: 10.1371/journal.pone.0139170. eCollection 2015. PLoS One. 2015. PMID: 26422397 Free PMC article.
-
Computational and experimental approaches for modeling gene regulatory networks.Curr Pharm Des. 2007;13(14):1415-36. doi: 10.2174/138161207780765945. Curr Pharm Des. 2007. PMID: 17504165 Review.
-
Chromatin organizer SATB1 as a novel molecular target for cancer therapy.Curr Drug Targets. 2012 Dec;13(13):1603-15. doi: 10.2174/138945012803530008. Curr Drug Targets. 2012. PMID: 22998183 Review.
Cited by
-
Distributed Bayesian networks reconstruction on the whole genome scale.PeerJ. 2018 Oct 19;6:e5692. doi: 10.7717/peerj.5692. eCollection 2018. PeerJ. 2018. PMID: 30364537 Free PMC article.
-
BNFinder2: Faster Bayesian network learning and Bayesian classification.Bioinformatics. 2013 Aug 15;29(16):2068-70. doi: 10.1093/bioinformatics/btt323. Epub 2013 Jul 1. Bioinformatics. 2013. PMID: 23818512 Free PMC article.
References
-
- Holter NS, Mitra M, Maritan A, Cieplak M, Banavar JR, Fedoroff NV. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8409–8414. doi: 10.1073/pnas.150242097. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

