Co-lethality studied as an asset against viral drug escape: the HIV protease case
- PMID: 20565756
- PMCID: PMC2898770
- DOI: 10.1186/1745-6150-5-40
Co-lethality studied as an asset against viral drug escape: the HIV protease case
Abstract
Background: Co-lethality, or synthetic lethality is the documented genetic situation where two, separately non-lethal mutations, become lethal when combined in one genome. Each mutation is called a "synthetic lethal" (SL) or a co-lethal. Like invariant positions, SL sets (SL linked couples) are choice targets for drug design against fast-escaping RNA viruses: mutational viral escape by loss of affinity to the drug may induce (synthetic) lethality.
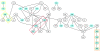
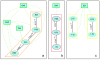
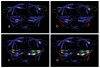
Results: From an amino acid sequence alignment of the HIV protease, we detected the potential SL couples, potential SL sets, and invariant positions. From the 3D structure of the same protein we focused on the ones that were close to each other and accessible on the protein surface, to possibly bind putative drugs. We aligned 24,155 HIV protease amino acid sequences and identified 290 potential SL couples and 25 invariant positions. After applying the distance and accessibility filter, three candidate drug design targets of respectively 7 (under the flap), 4 (in the cantilever) and 5 (in the fulcrum) amino acid positions were found.
Conclusions: These three replication-critical targets, located outside of the active site, are key to our anti-escape strategy. Indeed, biological evidence shows that 2/3 of those target positions perform essential biological functions. Their mutational variations to escape antiviral medication could be lethal, thus limiting the apparition of drug-resistant strains.
Reviewers: This article was reviewed by Arcady Mushegian, Shamil Sunyaev and Claus Wilke.
Figures
Similar articles
-
Synthetic lethals in HIV: ways to avoid drug resistance : Running title: Preventing HIV resistance.Biol Direct. 2015 Apr 17;10:17. doi: 10.1186/s13062-015-0044-y. Biol Direct. 2015. PMID: 25888435 Free PMC article.
-
Protein promiscuity: drug resistance and native functions--HIV-1 case.J Biomol Struct Dyn. 2005 Jun;22(6):615-24. doi: 10.1080/07391102.2005.10531228. J Biomol Struct Dyn. 2005. PMID: 15842167
-
Drug Resistance Mechanism of L10F, L10F/N88S and L90M mutations in CRF01_AE HIV-1 protease: Molecular dynamics simulations and binding free energy calculations.J Mol Graph Model. 2017 Aug;75:390-402. doi: 10.1016/j.jmgm.2017.06.007. Epub 2017 Jun 8. J Mol Graph Model. 2017. PMID: 28645089
-
Structural and thermodynamic basis of resistance to HIV-1 protease inhibition: implications for inhibitor design.Curr Drug Targets Infect Disord. 2003 Dec;3(4):311-28. doi: 10.2174/1568005033481051. Curr Drug Targets Infect Disord. 2003. PMID: 14754432 Review.
-
Adaptive inhibitors of the HIV-1 protease.Prog Biophys Mol Biol. 2005 Jun;88(2):193-208. doi: 10.1016/j.pbiomolbio.2004.07.005. Prog Biophys Mol Biol. 2005. PMID: 15572155 Review.
Cited by
-
Antitarget, Anti-SARS-CoV-2 Leads, Drugs, and the Drug Discovery-Genetics Alliance Perspective.J Med Chem. 2023 Mar 23;66(6):3664-3702. doi: 10.1021/acs.jmedchem.2c01229. Epub 2023 Mar 1. J Med Chem. 2023. PMID: 36857133 Free PMC article. Review.
-
VIRAPOPS2 supports the influenza virus reassortments.Source Code Biol Med. 2014 Aug 17;9:18. doi: 10.1186/1751-0473-9-18. eCollection 2014. Source Code Biol Med. 2014. PMID: 25183993 Free PMC article.
-
Synthetic lethals in HIV: ways to avoid drug resistance : Running title: Preventing HIV resistance.Biol Direct. 2015 Apr 17;10:17. doi: 10.1186/s13062-015-0044-y. Biol Direct. 2015. PMID: 25888435 Free PMC article.
-
A New Strategy to Reduce Influenza Escape: Detecting Therapeutic Targets Constituted of Invariance Groups.Viruses. 2017 Mar 2;9(3):38. doi: 10.3390/v9030038. Viruses. 2017. PMID: 28257108 Free PMC article.
References
-
- Noronha G, Cao J, Chow C, Dneprovskaia E, Hwang L, Lohse D, Ching Mak C, McPherson A, Fine RM, Kang X, Klebansky B, Palanki MSS, Pathak VP, Renick J, Soll R, Zeng B. Targeting drug resistant mutations using novel binding interactions - Lessons learned from Abl-T315I and their implications in drug design. Frontiers in drug design & discovery. 2007;3:121–144.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

