A general framework for optimization of probes for gene expression microarray and its application to the fungus Podospora anserina
- PMID: 20565839
- PMCID: PMC2908635
- DOI: 10.1186/1756-0500-3-171
A general framework for optimization of probes for gene expression microarray and its application to the fungus Podospora anserina
Abstract
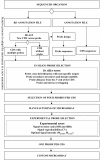
Background: The development of new microarray technologies makes custom long oligonucleotide arrays affordable for many experimental applications, notably gene expression analyses. Reliable results depend on probe design quality and selection. Probe design strategy should cope with the limited accuracy of de novo gene prediction programs, and annotation up-dating. We present a novel in silico procedure which addresses these issues and includes experimental screening, as an empirical approach is the best strategy to identify optimal probes in the in silico outcome.
Findings: We used four criteria for in silico probe selection: cross-hybridization, hairpin stability, probe location relative to coding sequence end and intron position. This latter criterion is critical when exon-intron gene structure predictions for intron-rich genes are inaccurate. For each coding sequence (CDS), we selected a sub-set of four probes. These probes were included in a test microarray, which was used to evaluate the hybridization behavior of each probe. The best probe for each CDS was selected according to three experimental criteria: signal-to-noise ratio, signal reproducibility, and representative signal intensities. This procedure was applied for the development of a gene expression Agilent platform for the filamentous fungus Podospora anserina and the selection of a single 60-mer probe for each of the 10,556 P. anserina CDS.
Conclusions: A reliable gene expression microarray version based on the Agilent 44K platform was developed with four spot replicates of each probe to increase statistical significance of analysis.
Figures



Similar articles
-
Development and validation of a custom microarray for global transcriptome profiling of the fungus Aspergillus nidulans.Curr Genet. 2016 Nov;62(4):897-910. doi: 10.1007/s00294-016-0597-z. Epub 2016 Apr 1. Curr Genet. 2016. PMID: 27038308
-
A revised design for microarray experiments to account for experimental noise and uncertainty of probe response.PLoS One. 2014 Mar 11;9(3):e91295. doi: 10.1371/journal.pone.0091295. eCollection 2014. PLoS One. 2014. PMID: 24618910 Free PMC article.
-
Empirical establishment of oligonucleotide probe design criteria.Appl Environ Microbiol. 2005 Jul;71(7):3753-60. doi: 10.1128/AEM.71.7.3753-3760.2005. Appl Environ Microbiol. 2005. PMID: 16000786 Free PMC article.
-
Calorie restriction in the filamentous fungus Podospora anserina.Exp Gerontol. 2010 Aug;45(7-8):516-24. doi: 10.1016/j.exger.2010.01.002. Epub 2010 Jan 11. Exp Gerontol. 2010. PMID: 20064602 Review.
-
The genome sequence of Podospora anserina, a classic model fungus.Genome Biol. 2008;9(5):223. doi: 10.1186/gb-2008-9-5-223. Epub 2008 May 15. Genome Biol. 2008. PMID: 18492226 Free PMC article. Review.
Cited by
-
Transcriptome analysis reveals unique metabolic features in the Cryptosporidium parvum Oocysts associated with environmental survival and stresses.BMC Genomics. 2012 Nov 21;13:647. doi: 10.1186/1471-2164-13-647. BMC Genomics. 2012. PMID: 23171372 Free PMC article.
-
Assessing De Novo transcriptome assembly metrics for consistency and utility.BMC Genomics. 2013 Jul 9;14:465. doi: 10.1186/1471-2164-14-465. BMC Genomics. 2013. PMID: 23837739 Free PMC article.
-
The transcriptional response to nonself in the fungus Podospora anserina.G3 (Bethesda). 2013 Jun 21;3(6):1015-30. doi: 10.1534/g3.113.006262. G3 (Bethesda). 2013. PMID: 23589521 Free PMC article.
-
A RID-like putative cytosine methyltransferase homologue controls sexual development in the fungus Podospora anserina.PLoS Genet. 2019 Aug 14;15(8):e1008086. doi: 10.1371/journal.pgen.1008086. eCollection 2019 Aug. PLoS Genet. 2019. PMID: 31412020 Free PMC article.
-
Genetic and functional investigation of Zn(2)Cys(6) transcription factors RSE2 and RSE3 in Podospora anserina.Eukaryot Cell. 2014 Jan;13(1):53-65. doi: 10.1128/EC.00172-13. Epub 2013 Nov 1. Eukaryot Cell. 2014. PMID: 24186951 Free PMC article.
References
-
- Paredes CJ, Senger RS, Spath IS, Borden JR, Sillers R, Papoutsakis ET. A general framework for designing and validating oligomer-based DNA microarrays and its application to Clostridium acetobutylicum. Applied and environmental microbiology. 2007;73:4631–4638. doi: 10.1128/AEM.00144-07. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

