Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns
- PMID: 20565864
- PMCID: PMC2917031
- DOI: 10.1186/bcr2590
Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns
Abstract
Introduction: Five different molecular subtypes of breast cancer have been identified through gene expression profiling. Each subtype has a characteristic expression pattern suggested to partly depend on cellular origin. We aimed to investigate whether the molecular subtypes also display distinct methylation profiles.
Methods: We analysed methylation status of 807 cancer-related genes in 189 fresh frozen primary breast tumours and four normal breast tissue samples using an array-based methylation assay.
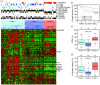
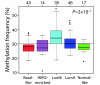
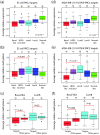
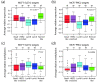
Results: Unsupervised analysis revealed three groups of breast cancer with characteristic methylation patterns. The three groups were associated with the luminal A, luminal B and basal-like molecular subtypes of breast cancer, respectively, whereas cancers of the HER2-enriched and normal-like subtypes were distributed among the three groups. The methylation frequencies were significantly different between subtypes, with luminal B and basal-like tumours being most and least frequently methylated, respectively. Moreover, targets of the polycomb repressor complex in breast cancer and embryonic stem cells were more methylated in luminal B tumours than in other tumours. BRCA2-mutated tumours had a particularly high degree of methylation. Finally, by utilizing gene expression data, we observed that a large fraction of genes reported as having subtype-specific expression patterns might be regulated through methylation.
Conclusions: We have found that breast cancers of the basal-like, luminal A and luminal B molecular subtypes harbour specific methylation profiles. Our results suggest that methylation may play an important role in the development of breast cancers.
Figures


References
-
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. - PubMed
-
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

