The CaaX specificities of Arabidopsis protein prenyltransferases explain era1 and ggb phenotypes
- PMID: 20565889
- PMCID: PMC3017772
- DOI: 10.1186/1471-2229-10-118
The CaaX specificities of Arabidopsis protein prenyltransferases explain era1 and ggb phenotypes
Abstract
Background: Protein prenylation is a common post-translational modification in metazoans, protozoans, fungi, and plants. This modification, which mediates protein-membrane and protein-protein interactions, is characterized by the covalent attachment of a fifteen-carbon farnesyl or twenty-carbon geranylgeranyl group to the cysteine residue of a carboxyl terminal CaaX motif. In Arabidopsis, era1 mutants lacking protein farnesyltransferase exhibit enlarged meristems, supernumerary floral organs, an enhanced response to abscisic acid (ABA), and drought tolerance. In contrast, ggb mutants lacking protein geranylgeranyltransferase type 1 exhibit subtle changes in ABA and auxin responsiveness, but develop normally.
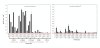
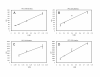
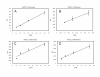
Results: We have expressed recombinant Arabidopsis protein farnesyltransferase (PFT) and protein geranylgeranyltransferase type 1 (PGGT1) in E. coli and characterized purified enzymes with respect to kinetic constants and substrate specificities. Our results indicate that, whereas PFT exhibits little specificity for the terminal amino acid of the CaaX motif, PGGT1 exclusively prenylates CaaX proteins with a leucine in the terminal position. Moreover, we found that different substrates exhibit similar K(m) but different k(cat) values in the presence of PFT and PGGT1, indicating that substrate specificities are determined primarily by reactivity rather than binding affinity.
Conclusions: The data presented here potentially explain the relatively strong phenotype of era1 mutants and weak phenotype of ggb mutants. Specifically, the substrate specificities of PFT and PGGT1 suggest that PFT can compensate for loss of PGGT1 in ggb mutants more effectively than PGGT1 can compensate for loss of PFT in era1 mutants. Moreover, our results indicate that PFT and PGGT1 substrate specificities are primarily due to differences in catalysis, rather than differences in substrate binding.
Figures




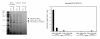
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

