Duplication, divergence and persistence in the Phytochrome photoreceptor gene family of cottons (Gossypium spp.)
- PMID: 20565911
- PMCID: PMC3095280
- DOI: 10.1186/1471-2229-10-119
Duplication, divergence and persistence in the Phytochrome photoreceptor gene family of cottons (Gossypium spp.)
Abstract
Background: Phytochromes are a family of red/far-red photoreceptors that regulate a number of important developmental traits in cotton (Gossypium spp.), including plant architecture, fiber development, and photoperiodic flowering. Little is known about the composition and evolution of the phytochrome gene family in diploid (G. herbaceum, G. raimondii) or allotetraploid (G. hirsutum, G. barbadense) cotton species. The objective of this study was to obtain a preliminary inventory and molecular-evolutionary characterization of the phytochrome gene family in cotton.
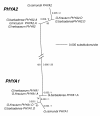
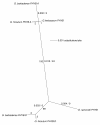
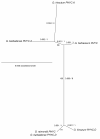
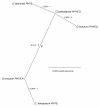
Results: We used comparative sequence resources to design low-degeneracy PCR primers that amplify genomic sequence tags (GSTs) for members of the PHYA, PHYB/D, PHYC and PHYE gene sub-families from A- and D-genome diploid and AD-genome allotetraploid Gossypium species. We identified two paralogous PHYA genes (designated PHYA1 and PHYA2) in diploid cottons, the result of a Malvaceae-specific PHYA gene duplication that occurred approximately 14 million years ago (MYA), before the divergence of the A- and D-genome ancestors. We identified a single gene copy of PHYB, PHYC, and PHYE in diploid cottons. The allotetraploid genomes have largely retained the complete gene complements inherited from both of the diploid genome ancestors, with at least four PHYA genes and two genes encoding PHYB, PHYC and PHYE in the AD-genomes. We did not identify a PHYD gene in any cotton genomes examined.
Conclusions: Detailed sequence analysis suggests that phytochrome genes retained after duplication by segmental duplication and allopolyploidy appear to be evolving independently under a birth-and-death-process with strong purifying selection. Our study provides a preliminary phytochrome gene inventory that is necessary and sufficient for further characterization of the biological functions of each of the cotton phytochrome genes, and for the development of 'candidate gene' markers that are potentially useful for cotton improvement via modern marker-assisted selection strategies.
Figures

Similar articles
-
Development, genetic mapping and QTL association of cotton PHYA, PHYB, and HY5-specific CAPS and dCAPS markers.BMC Genet. 2016 Oct 24;17(1):141. doi: 10.1186/s12863-016-0448-4. BMC Genet. 2016. PMID: 27776497 Free PMC article.
-
Genome-Wide Comparative Analysis of the Phospholipase D Gene Families among Allotetraploid Cotton and Its Diploid Progenitors.PLoS One. 2016 May 23;11(5):e0156281. doi: 10.1371/journal.pone.0156281. eCollection 2016. PLoS One. 2016. PMID: 27213891 Free PMC article.
-
Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species.BMC Genomics. 2017 Apr 12;18(1):292. doi: 10.1186/s12864-017-3682-x. BMC Genomics. 2017. PMID: 28403834 Free PMC article.
-
Evolutionary studies illuminate the structural-functional model of plant phytochromes.Plant Cell. 2010 Jan;22(1):4-16. doi: 10.1105/tpc.109.072280. Epub 2010 Jan 29. Plant Cell. 2010. PMID: 20118225 Free PMC article. Review.
-
The Current Progresses in the Genes and Networks Regulating Cotton Plant Architecture.Front Plant Sci. 2022 Jun 9;13:882583. doi: 10.3389/fpls.2022.882583. eCollection 2022. Front Plant Sci. 2022. PMID: 35755647 Free PMC article. Review.
Cited by
-
Development, genetic mapping and QTL association of cotton PHYA, PHYB, and HY5-specific CAPS and dCAPS markers.BMC Genet. 2016 Oct 24;17(1):141. doi: 10.1186/s12863-016-0448-4. BMC Genet. 2016. PMID: 27776497 Free PMC article.
-
Ectopic expression reveals a conserved PHYB homolog in soybean.PLoS One. 2011;6(11):e27737. doi: 10.1371/journal.pone.0027737. Epub 2011 Nov 16. PLoS One. 2011. PMID: 22110748 Free PMC article.
-
Gene flow signature in the S-allele region of cultivated buckwheat.BMC Plant Biol. 2019 Apr 3;19(1):125. doi: 10.1186/s12870-019-1730-1. BMC Plant Biol. 2019. PMID: 30943914 Free PMC article.
-
The phytochrome gene family in soybean and a dominant negative effect of a soybean PHYA transgene on endogenous Arabidopsis PHYA.Plant Cell Rep. 2013 Dec;32(12):1879-90. doi: 10.1007/s00299-013-1500-8. Epub 2013 Sep 8. Plant Cell Rep. 2013. PMID: 24013793
-
Functional dissection of phytochrome A in plants.Front Plant Sci. 2024 Jan 26;15:1340260. doi: 10.3389/fpls.2024.1340260. eCollection 2024. Front Plant Sci. 2024. PMID: 38344182 Free PMC article. Review.
References
-
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in plants. 2. Kluwer Academic Publishers, Dordrecht, the Netherlands; 1994.
-
- Smith H. Physiological and Ecological Function within the Phytochrome Family. Annu Rev Plant Phys. 1995;46:289–315. doi: 10.1146/annurev.pp.46.060195.001445. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

