A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum
- PMID: 20565933
- PMCID: PMC3055265
- DOI: 10.1186/1471-2148-10-191
A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum
Abstract
Background: Plastid replacements through secondary endosymbioses include massive transfer of genes from the endosymbiont to the host nucleus and require a new targeting system to enable transport of the plastid-targeted proteins across 3-4 plastid membranes. The dinoflagellates are the only eukaryotic lineage that has been shown to have undergone several plastid replacement events, and this group is thus highly relevant for studying the processes involved in plastid evolution. In this study, we analyzed the phylogenetic origin and N-terminal extensions of plastid-targeted proteins from Lepidodinium chlorophorum, a member of the only dinoflagellate genus that harbors a green secondary plastid rather than the red algal-derived, peridinin-containing plastid usually found in photosynthetic dinoflagellates.
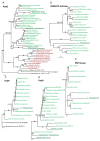
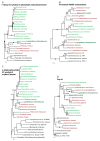
Results: We sequenced 4,746 randomly picked clones from a L. chlorophorum cDNA library. 22 of the assembled genes were identified as genes encoding proteins functioning in plastids. Some of these were of green algal origin. This confirms that genes have been transferred from the plastid to the host nucleus of L. chlorophorum and indicates that the plastid is fully integrated as an organelle in the host. Other nuclear-encoded plastid-targeted protein genes, however, are clearly not of green algal origin, but have been derived from a number of different algal groups, including dinoflagellates, streptophytes, heterokonts, and red algae. The characteristics of N-terminal plastid-targeting peptides of all of these genes are substantially different from those found in peridinin-containing dinoflagellates and green algae.
Conclusions: L. chlorophorum expresses plastid-targeted proteins with a range of different origins, which probably arose through endosymbiotic gene transfer (EGT) and horizontal gene transfer (HGT). The N-terminal extension of the genes is different from the extensions found in green alga and other dinoflagellates (peridinin- and haptophyte plastids). These modifications have likely enabled the mosaic proteome of L. chlorophorum.
Figures

Similar articles
-
Plastid genome-based phylogeny pinpointed the origin of the green-colored plastid in the dinoflagellate Lepidodinium chlorophorum.Genome Biol Evol. 2015 Apr 2;7(4):1133-40. doi: 10.1093/gbe/evv060. Genome Biol Evol. 2015. PMID: 25840416 Free PMC article.
-
Origins of plastids and glyceraldehyde-3-phosphate dehydrogenase genes in the green-colored dinoflagellate Lepidodinium chlorophorum.Gene. 2008 Feb 29;410(1):26-36. doi: 10.1016/j.gene.2007.11.008. Epub 2007 Nov 28. Gene. 2008. PMID: 18191504
-
Prasinoxanthin is absent in the green-colored dinoflagellate Lepidodinium chlorophorum strain NIES-1868: pigment composition and 18S rRNA phylogeny.J Plant Res. 2012 Nov;125(6):705-11. doi: 10.1007/s10265-012-0486-6. Epub 2012 Mar 23. J Plant Res. 2012. PMID: 22441568
-
The endosymbiotic origin, diversification and fate of plastids.Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):729-48. doi: 10.1098/rstb.2009.0103. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124341 Free PMC article. Review.
-
Plastids and protein targeting.J Eukaryot Microbiol. 1999 Jul-Aug;46(4):339-46. doi: 10.1111/j.1550-7408.1999.tb04613.x. J Eukaryot Microbiol. 1999. PMID: 10461382 Review.
Cited by
-
Plastid origin and evolution: new models provide insights into old problems.Plant Physiol. 2011 Apr;155(4):1552-60. doi: 10.1104/pp.111.173500. Epub 2011 Feb 22. Plant Physiol. 2011. PMID: 21343425 Free PMC article. Review. No abstract available.
-
Integration of plastids with their hosts: Lessons learned from dinoflagellates.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10247-54. doi: 10.1073/pnas.1421380112. Epub 2015 May 20. Proc Natl Acad Sci U S A. 2015. PMID: 25995366 Free PMC article. Review.
-
Genomic perspectives on the birth and spread of plastids.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10147-53. doi: 10.1073/pnas.1421374112. Epub 2015 Apr 20. Proc Natl Acad Sci U S A. 2015. PMID: 25902528 Free PMC article.
-
Comparative Plastid Genomics of Green-Colored Dinoflagellates Unveils Parallel Genome Compaction and RNA Editing.Front Plant Sci. 2022 Jul 11;13:918543. doi: 10.3389/fpls.2022.918543. eCollection 2022. Front Plant Sci. 2022. PMID: 35898209 Free PMC article.
-
Secondary Plastids of Euglenids and Chlorarachniophytes Function with a Mix of Genes of Red and Green Algal Ancestry.Mol Biol Evol. 2018 Sep 1;35(9):2198-2204. doi: 10.1093/molbev/msy121. Mol Biol Evol. 2018. PMID: 29924337 Free PMC article.
References
-
- Gray MW, Spencer DF. In: Evolution of Microbial Life. Roberts DM, Sharp P, Alderson G, editor. Vol. 54. Collins MA Cambridge: Cambridge University Press; 1996. Organellar evolution; pp. 109–126.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

