Array comparative hybridisation reveals a high degree of similarity between UK and European clinical isolates of hypervirulent Clostridium difficile
- PMID: 20565959
- PMCID: PMC3224701
- DOI: 10.1186/1471-2164-11-389
Array comparative hybridisation reveals a high degree of similarity between UK and European clinical isolates of hypervirulent Clostridium difficile
Abstract
Background: Clostridium difficile is a Gram-positive, anaerobic, spore-forming bacterium that is responsible for C. difficile associated disease in humans and is currently the most common cause of nosocomial diarrhoea in the western world. This current status has been linked to the emergence of a highly virulent PCR-ribotype 027 strain. The aim of this work was to identify regions of sequence divergence that may be used as genetic markers of hypervirulent PCR-ribotype 027 strains and markers of the sequenced strain, CD630 by array comparative hybridisation.
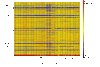
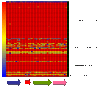
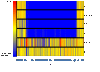
Results: In this study, we examined 94 clinical strains of the most common PCR-ribotypes isolated in mainland Europe and the UK by array comparative genomic hybridisation. Our array was comprehensive with 40,097 oligonucleotides covering the C. difficile 630 genome and revealed a core genome for all the strains of 32%. The array also covered genes unique to two PCR-ribotype 027 strains, relative to C. difficile 630 which were represented by 681 probes. All of these genes were also found in the commonly occuring PCR-ribotypes 001 and 106, and the emerging hypervirulent PCR-ribotype 078 strains, indicating that these are markers for all highly virulent strains.
Conclusions: We have fulfilled the aims of this study by identifying markers for CD630 and markers associated with hypervirulence, albeit genes that are not just indicative of PCR-ribotype 027 strains. We have also extended this study and have defined a more stringent core gene set compared to those previously published due to the comprehensive array coverage. Further to this we have defined a list of genes absent from non-toxinogenic strains and defined the nature of the specific toxin deletion in the strain CD37.
Figures
Similar articles
-
Sequence similarity of Clostridium difficile strains by analysis of conserved genes and genome content is reflected by their ribotype affiliation.PLoS One. 2014 Jan 23;9(1):e86535. doi: 10.1371/journal.pone.0086535. eCollection 2014. PLoS One. 2014. PMID: 24482682 Free PMC article.
-
Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains.J Bacteriol. 2006 Oct;188(20):7297-305. doi: 10.1128/JB.00664-06. J Bacteriol. 2006. PMID: 17015669 Free PMC article.
-
Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium.Genome Biol. 2009;10(9):R102. doi: 10.1186/gb-2009-10-9-r102. Epub 2009 Sep 25. Genome Biol. 2009. PMID: 19781061 Free PMC article.
-
The Clostridium difficile PCR ribotype 027 lineage: a pathogen on the move.Clin Microbiol Infect. 2014 May;20(5):396-404. doi: 10.1111/1469-0691.12619. Epub 2014 Apr 28. Clin Microbiol Infect. 2014. PMID: 24621128 Review.
-
The emergence of 'hypervirulence' in Clostridium difficile.Int J Med Microbiol. 2010 Aug;300(6):387-95. doi: 10.1016/j.ijmm.2010.04.008. Epub 2010 May 23. Int J Med Microbiol. 2010. PMID: 20547099 Review.
Cited by
-
Comparative transcriptional analysis of clinically relevant heat stress response in Clostridium difficile strain 630.PLoS One. 2012;7(7):e42410. doi: 10.1371/journal.pone.0042410. Epub 2012 Jul 30. PLoS One. 2012. PMID: 22860125 Free PMC article.
-
The agr locus regulates virulence and colonization genes in Clostridium difficile 027.J Bacteriol. 2013 Aug;195(16):3672-81. doi: 10.1128/JB.00473-13. Epub 2013 Jun 14. J Bacteriol. 2013. PMID: 23772065 Free PMC article.
-
Clostridioides difficile ribotype 106: A systematic review of the antimicrobial susceptibility, genetics, and clinical outcomes of this common worldwide strain.Anaerobe. 2020 Apr;62:102142. doi: 10.1016/j.anaerobe.2019.102142. Epub 2019 Dec 19. Anaerobe. 2020. PMID: 32007682 Free PMC article.
-
Analysis of a Clostridium difficile PCR ribotype 078 100 kilobase island reveals the presence of a novel transposon, Tn6164.BMC Microbiol. 2012 Jul 2;12:130. doi: 10.1186/1471-2180-12-130. BMC Microbiol. 2012. PMID: 22747711 Free PMC article.
-
Extended multilocus variable-number tandem-repeat analysis of Clostridium difficile correlates exactly with ribotyping and enables identification of hospital transmission.J Clin Microbiol. 2011 Oct;49(10):3523-30. doi: 10.1128/JCM.00546-11. Epub 2011 Aug 17. J Clin Microbiol. 2011. PMID: 21849691 Free PMC article.
References
-
- Elixhauser A, Jhung M. HCUP statistical brief No. 50. US Agency for Healthcare Research and Quality, Rockville, MD; 2008. Clostridium difficile-associated disease in US hospitals, 1993-2005. - PubMed
-
- Barbut F, Gariazzo B, Bonne L, Lalande V, Burghoffer B, Luiuz R, Petit JC. Clinical features of Clostridium difficile-associated infections and molecular characterization of strains: results of a retrospective study, 2000-2004. Infect Control Hosp Epidemiol. 2007;28:131–139. doi: 10.1086/511794. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

