Genome comparison of the epiphytic bacteria Erwinia billingiae and E. tasmaniensis with the pear pathogen E. pyrifoliae
- PMID: 20565991
- PMCID: PMC2897811
- DOI: 10.1186/1471-2164-11-393
Genome comparison of the epiphytic bacteria Erwinia billingiae and E. tasmaniensis with the pear pathogen E. pyrifoliae
Abstract
Background: The genus Erwinia includes plant-associated pathogenic and non-pathogenic Enterobacteria. Important pathogens such as Erwinia amylovora, the causative agent of fire blight and E. pyrifoliae causing bacterial shoot blight of pear in Asia belong to this genus. The species E. tasmaniensis and E. billingiae are epiphytic bacteria and may represent antagonists for biocontrol of fire blight. The presence of genes that are putatively involved in virulence in E. amylovora and E. pyrifoliae is of special interest for these species in consequence.
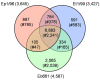
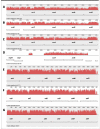
Results: Here we provide the complete genome sequences of the pathogenic E. pyrifoliae strain Ep1/96 with a size of 4.1 Mb and of the non-pathogenic species E. billingiae strain Eb661 with a size of 5.4 Mb, de novo determined by conventional Sanger sequencing and next generation sequencing techniques. Genome comparison reveals large inversions resulting from homologous recombination events. Furthermore, comparison of deduced proteins highlights a relation of E. billingiae strain Eb661 to E. tasmaniensis strain Et1/99 and a distance to E. pyrifoliae for the overall gene content as well as for the presence of encoded proteins representing virulence factors for the pathogenic species. Pathogenicity of E. pyrifoliae is supposed to have evolved by accumulation of potential virulence factors. E. pyrifoliae carries factors for type III secretion and cell invasion. Other genes described as virulence factors for E. amylovora are involved in the production of exopolysaccharides, the utilization of plant metabolites such as sorbitol and sucrose. Some virulence-associated genes of the pathogenic species are present in E. tasmaniensis but mostly absent in E. billingiae.
Conclusion: The data of the genome analyses correspond to the pathogenic lifestyle of E. pyrifoliae and underlines the epiphytic localization of E. tasmaniensis and E. billingiae as a saprophyte.
Figures

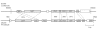

References
-
- Rhim SL, Volksch B, Gardan L, Paulin JP, Langlotz C, Kim WS, Geider K. Erwinia pyrifoliae, an Erwinia species different from Erwinia amylovora, causes a necrotic disease of Asian pear trees. Plant Pathology. 1999;48(4):514–520. doi: 10.1046/j.1365-3059.1999.00376.x. - DOI
-
- Shrestha R, Koo J, Park D, Hwang I, Hur J, Lim C. Erwinia pyrifoliae, a causal endemic pathogen of shoot blight of Asian pear tree in Korea. The Plant Pathology Journal. 2003;19(6):294–300.
-
- Vanneste J. Fire blight: The disease and its causative agent, Erwinia amylovora. Wallingford, Oxon, UK; New York, NY, USA: CABI Pub; 2000. p. 370.
-
- Loper JE, Henkels MD, Roberts RG, Grove GG, Willett MJ, Smith TJ. Evaluation of Streptomycin, Oxytetracycline, and Copper Resistance of Erwinia amylovora isolated from Pear Orchards in Washington-State. Plant Disease. 1991;75(3):287–290.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

