Identification of a substrate-binding site in a peroxisomal beta-oxidation enzyme by photoaffinity labeling with a novel palmitoyl derivative
- PMID: 20566640
- PMCID: PMC2924054
- DOI: 10.1074/jbc.M110.104547
Identification of a substrate-binding site in a peroxisomal beta-oxidation enzyme by photoaffinity labeling with a novel palmitoyl derivative
Abstract
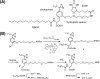
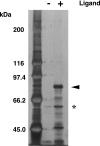
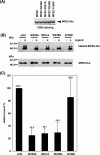
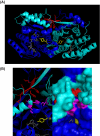
Peroxisomes play an essential role in a number of important metabolic pathways including beta-oxidation of fatty acids and their derivatives. Therefore, peroxisomes possess various beta-oxidation enzymes and specialized fatty acid transport systems. However, the molecular mechanisms of these proteins, especially in terms of substrate binding, are still unknown. In this study, to identify the substrate-binding sites of these proteins, we synthesized a photoreactive palmitic acid analogue bearing a diazirine moiety as a photophore, and performed photoaffinity labeling of purified rat liver peroxisomes. As a result, an 80-kDa peroxisomal protein was specifically labeled by the photoaffinity ligand, and the labeling efficiency competitively decreased in the presence of palmitoyl-CoA. Mass spectrometric analysis identified the 80-kDa protein as peroxisomal multifunctional enzyme type 2 (MFE2), one of the peroxisomal beta-oxidation enzymes. Recombinant rat MFE2 was also labeled by the photoaffinity ligand, and mass spectrometric analysis revealed that a fragment of rat MFE2 (residues Trp(249) to Arg(251)) was labeled by the ligand. MFE2 mutants bearing these residues, MFE2(W249A) and MFE2(R251A), exhibited decreased labeling efficiency. Furthermore, MFE2(W249G), which corresponds to one of the disease-causing mutations in human MFE2, also exhibited a decreased efficiency. Based on the crystal structure of rat MFE2, these residues are located on the top of a hydrophobic cavity leading to an active site of MFE2. These data suggest that MFE2 anchors its substrate around the region from Trp(249) to Arg(251) and positions the substrate along the hydrophobic cavity in the proper direction toward the catalytic center.
Figures





References
-
- Wanders R. J., Waterham H. R. (2006) Annu. Rev. Biochem. 75, 295–332 - PubMed
-
- Lazarow P. B., Moser H. W. (1995) The Metabolic and Molecular Basis of Inherited Disease, pp. 2287–2324, McGraw-Hill Inc., New York
-
- Fujiki Y. (2000) FEBS Lett. 476, 42–46 - PubMed
-
- Wanders R. J., Waterham H. R. (2006) Biochim. Biophys. Acta 1763, 1707–1720 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

