Dual recognition of CENP-A nucleosomes is required for centromere assembly
- PMID: 20566683
- PMCID: PMC2894454
- DOI: 10.1083/jcb.201001013
Dual recognition of CENP-A nucleosomes is required for centromere assembly
Abstract
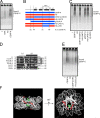
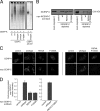
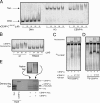
Centromeres contain specialized nucleosomes in which histone H3 is replaced by the histone variant centromere protein A (CENP-A). CENP-A nucleosomes are thought to act as an epigenetic mark that specifies centromere identity. We previously identified CENP-N as a CENP-A nucleosome-specific binding protein. Here, we show that CENP-C also binds directly and specifically to CENP-A nucleosomes. Nucleosome binding by CENP-C required the extreme C terminus of CENP-A and did not compete with CENP-N binding, which suggests that CENP-C and CENP-N recognize distinct structural elements of CENP-A nucleosomes. A mutation that disrupted CENP-C binding to CENP-A nucleosomes in vitro caused defects in CENP-C targeting to centromeres. Moreover, depletion of CENP-C with siRNA resulted in the mislocalization of all other nonhistone CENPs examined, including CENP-K, CENP-H, CENP-I, and CENP-T, and led to a partial reduction in centromeric CENP-A. We propose that CENP-C binds directly to CENP-A chromatin and, together with CENP-N, provides the foundation upon which other centromere and kinetochore proteins are assembled.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

