Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence
- PMID: 20566694
- PMCID: PMC2937430
- DOI: 10.1128/IAI.00165-10
Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence
Abstract
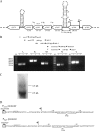
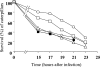
Methionine sulfoxide reductases A and B are antioxidant repair enzymes that reduce the S- and R-diastereomers of methionine sulfoxides back to methionine, respectively. Enterococcus faecalis, an important nosocomial pathogen, has one msrA gene and one msrB gene situated in different parts of the chromosome. Promoters have been mapped and mutants have been constructed in two E. faecalis strains (strains JH2-2 and V583) and characterized. For both backgrounds, the mutants are more sensitive than the wild-type parents to exposure to H2O2, and in combination the mutations seem to be additive. The virulence of the mutants has been analyzed in four different models. Survival of the mutants inside mouse peritoneal macrophages stimulated with recombinant gamma interferon plus lipopolysaccharide but not in naïve phagocytes is significantly affected. The msrA mutant is attenuated in the Galleria mellonella insect model. Deficiency in either Msr enzyme reduced the level of virulence in a systemic and urinary tract infection model. Virulence was reconstituted in the complemented strains. The combined results show that Msr repair enzymes are important for the oxidative stress response, macrophage survival, and persistent infection with E. faecalis.
Figures



References
-
- Alamuri, P., and R. J. Maier. 2004. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:1397-1406. - PubMed
-
- Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. J. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. - PubMed
-
- Benachour, A., Y. Auffray, and A. Hartke. 2007. Construction of plasmid vectors for screening replicons from gram-positive bacteria and their use as shuttle cloning vectors. Curr. Microbiol. 54:342-347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

