In vitro reconstitution of PHO5 promoter chromatin remodeling points to a role for activator-nucleosome competition in vivo
- PMID: 20566699
- PMCID: PMC2916437
- DOI: 10.1128/MCB.01399-09
In vitro reconstitution of PHO5 promoter chromatin remodeling points to a role for activator-nucleosome competition in vivo
Abstract
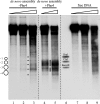
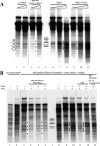
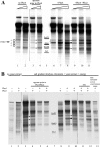
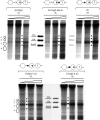
The yeast PHO5 promoter is a classical model for studying the role of chromatin in gene regulation. To enable biochemical dissection of the mechanism leading to PHO5 activation, we reconstituted the process in vitro. Positioned nucleosomes corresponding to the repressed PHO5 promoter state were assembled using a yeast extract-based in vitro system. Addition of the transactivator Pho4 yielded an extensive DNase I-hypersensitive site resembling induced PHO5 promoter chromatin. Importantly, this remodeling was energy dependent. In contrast, little or no chromatin remodeling was detected at the PHO8 or PHO84 promoter in this in vitro system. Only the PHO5 promoter harbors a high-affinity intranucleosomal Pho4 binding site (UASp) where Pho4 binding can compete with nucleosome formation, prompting us to test the importance of such competition for chromatin remodeling by analysis of UASp mutants in vivo. Indeed, the intranucleosomal location of the UASp element was critical, but not essential, for complete remodeling at the PHO5 promoter in vivo. Further, binding of just the Gal4 DNA binding domain to an intranucleosomal site could increase PHO5 promoter opening. These data establish an auxiliary role for DNA binding competition between Pho4 and histones in PHO5 promoter chromatin remodeling in vivo.
Figures





Similar articles
-
Effective dynamics of nucleosome configurations at the yeast PHO5 promoter.Elife. 2021 Mar 5;10:e58394. doi: 10.7554/eLife.58394. Elife. 2021. PMID: 33666171 Free PMC article.
-
The yeast PHO5 promoter: from single locus to systems biology of a paradigm for gene regulation through chromatin.Nucleic Acids Res. 2014;42(17):10888-902. doi: 10.1093/nar/gku784. Epub 2014 Sep 4. Nucleic Acids Res. 2014. PMID: 25190457 Free PMC article. Review.
-
Differential cofactor requirements for histone eviction from two nucleosomes at the yeast PHO84 promoter are determined by intrinsic nucleosome stability.Mol Cell Biol. 2009 Jun;29(11):2960-81. doi: 10.1128/MCB.01054-08. Epub 2009 Mar 23. Mol Cell Biol. 2009. PMID: 19307305 Free PMC article.
-
Transcriptional regulation of the yeast PHO8 promoter in comparison to the coregulated PHO5 promoter.J Biol Chem. 2000 Jul 28;275(30):22678-85. doi: 10.1074/jbc.M001409200. J Biol Chem. 2000. PMID: 10801809
-
Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast.Trends Biochem Sci. 1997 Mar;22(3):93-7. doi: 10.1016/s0968-0004(97)01001-3. Trends Biochem Sci. 1997. PMID: 9066259 Review.
Cited by
-
Effective dynamics of nucleosome configurations at the yeast PHO5 promoter.Elife. 2021 Mar 5;10:e58394. doi: 10.7554/eLife.58394. Elife. 2021. PMID: 33666171 Free PMC article.
-
The RSC chromatin remodeling complex has a crucial role in the complete remodeler set for yeast PHO5 promoter opening.Nucleic Acids Res. 2014 Apr;42(7):4270-82. doi: 10.1093/nar/gkt1395. Epub 2014 Jan 24. Nucleic Acids Res. 2014. PMID: 24465003 Free PMC article.
-
DNA sequence preferences of transcriptional activators correlate more strongly than repressors with nucleosomes.Mol Cell. 2012 Jul 27;47(2):183-92. doi: 10.1016/j.molcel.2012.06.028. Mol Cell. 2012. PMID: 22841002 Free PMC article.
-
The yeast PHO5 promoter: from single locus to systems biology of a paradigm for gene regulation through chromatin.Nucleic Acids Res. 2014;42(17):10888-902. doi: 10.1093/nar/gku784. Epub 2014 Sep 4. Nucleic Acids Res. 2014. PMID: 25190457 Free PMC article. Review.
-
Role of transcription factor-mediated nucleosome disassembly in PHO5 gene expression.Sci Rep. 2016 Feb 4;6:20319. doi: 10.1038/srep20319. Sci Rep. 2016. PMID: 26843321 Free PMC article.
References
-
- Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. - PubMed
-
- Barbarić, S., T. Luckenbach, A. Schmid, D. Blaschke, W. Hörz, and P. Korber. 2007. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J. Biol. Chem. 282:27610-27621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
