DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously
- PMID: 20566706
- PMCID: PMC2923886
- DOI: 10.1104/pp.110.156943
DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously
Abstract
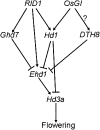
The three most important agronomic traits of rice (Oryza sativa), yield, plant height, and flowering time, are controlled by many quantitative trait loci (QTLs). In this study, a newly identified QTL, DTH8 (QTL for days to heading on chromosome 8), was found to regulate these three traits in rice. Map-based cloning reveals that DTH8 encodes a putative HAP3 subunit of the CCAAT-box-binding transcription factor and the complementary experiment increased significantly days to heading, plant height, and number of grains per panicle in CSSL61 (a chromosome segment substitution line that carries the nonfunctional DTH8 allele) with the Asominori functional DTH8 allele under long-day conditions. DTH8 is expressed in most tissues and its protein is localized to the nucleus exclusively. The quantitative real-time PCR assay revealed that DTH8 could down-regulate the transcriptions of Ehd1 (for Early heading date1) and Hd3a (for Heading date3a; a rice ortholog of FLOWERING LOCUS T) under long-day conditions. Ehd1 and Hd3a can also be down-regulated by the photoperiodic flowering genes Ghd7 and Hd1 (a rice ortholog of CONSTANS). Meanwhile, the transcription of DTH8 has been proved to be independent of Ghd7 and Hd1, and the natural mutation of this gene caused weak photoperiod sensitivity and shorter plant height. Taken together, these data indicate that DTH8 probably plays an important role in the signal network of photoperiodic flowering as a novel suppressor as well as in the regulation of plant height and yield potential.
Figures




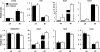



References
-
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 - PubMed
-
- Ashikari M, Sakakibara H, Lin SY, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. (2005) Cytokinin oxidase regulates rice grain production. Science 309: 741–745 - PubMed
-
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

