Parvovirus B19 genotype specific amino acid substitution in NS1 reduces the protein's cytotoxicity in culture
- PMID: 20567611
- PMCID: PMC2880839
- DOI: 10.7150/ijms.7.110
Parvovirus B19 genotype specific amino acid substitution in NS1 reduces the protein's cytotoxicity in culture
Abstract
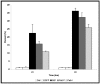
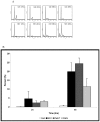
A clinical association between idiopathic liver disease and parvovirus B19 infection has been observed. Fulminant liver failure, not associated with other liver-tropic viruses, has been attributed to B19 in numerous reports, suggesting a possible role for B19 components in the extensive hepatocyte cytotoxicity observed in this condition. A recent report by Abe and colleagues (Int J Med Sci. 2007;4:105-9) demonstrated a link between persistent parvovirus B19 genotype I and III infection and fulminant liver failure. The genetic analysis of isolates obtained from these patients demonstrated a conservation of key amino acids in the nonstructural protein 1 (NS1) of the disease-associated genotypes. In this report we examine a conserved residue identified by Abe and colleagues and show that substitution of isoleucine 181 for methionine, as occurs in B19 genotype II, results in the reduction of B19 NS1-induced cytotoxicity of liver cells. Our results support the hypothesis that in the setting of persistent B19 infection, direct B19 NS1-induced cytotoxicity may play a role in idiopathic fulminant liver failure.
Keywords: Apoptosis; B19; Cytotoxicity; Fulminant Liver Failure; Parvovirus.
Conflict of interest statement
Conflict of Interest: The authors have declared that no conflict of interest exists.
Figures
References
-
- Anderson M.J, Higgins PG, Davis LR, Willman JS, Jones SE, Kidd IM, Pattison JR, Tyrrell DA. Experimental parvoviral infection in humans. J Infect Dis. 1985;152:257–65. - PubMed
-
- Bonvicini F, Filippone C, Manaresi E, Zerbini M, Musiani M, Gallinella G. HepG2 hepatocellular carcinoma cells are a non-permissive system for B19 virus infection. J Gen Virol. 2008;89:3034–8. - PubMed
-
- Brown K.E, Anderson SM, Young NS. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–7. - PubMed
-
- Brunstein J, Soderlund-Venermo M, Hedman K. Identification of a novel RNA splicing pattern as a basis of restricted cell tropism of erythrovirus B19. Virology. 2000;274:284–91. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

