Evidence, from simulations, of a single state with residual native structure at the thermal denaturation midpoint of a small globular protein
- PMID: 20568747
- PMCID: PMC2910365
- DOI: 10.1021/ja1031503
Evidence, from simulations, of a single state with residual native structure at the thermal denaturation midpoint of a small globular protein
Abstract
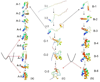
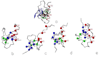
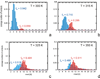
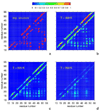
The folding of the B-domain of staphylococcal protein A has been studied by coarse-grained canonical and multiplexed replica-exchange molecular dynamics simulations with the UNRES force field in a broad range of temperatures (270 K < or = T < or = 350 K). In canonical simulations, the folding was found to occur either directly to the native state or through kinetic traps, mainly the topological mirror image of the native three-helix bundle. The latter folding scenario was observed more frequently at low temperatures. With increase of temperature, the frequency of the transitions between the folded and misfolded/unfolded states increased and the folded state became more diffuse with conformations exhibiting increased root-mean-square deviations from the experimental structure (from about 4 A at T = 300 K to 8.7 A at T = 325 K). An analysis of the equilibrium conformational ensemble determined from multiplexed replica exchange simulations at the folding-transition temperature (T(f) = 325 K) showed that the conformational ensemble at this temperature is a collection of conformations with residual secondary structures, which possess native or near-native clusters of nonpolar residues in place, and not a 50-50% mixture of fully folded and fully unfolded conformations. These findings contradict the quasi-chemical picture of two- or multistate protein folding, which assumes an equilibrium between the folded, unfolded, and intermediate states, with equilibrium shifting with temperature but with the native conformations remaining essentially unchanged. Our results also suggest that long-range hydrophobic contacts are the essential factor to keep the structure of a protein thermally stable.
Figures





Similar articles
-
Kinetic studies of folding of the B-domain of staphylococcal protein A with molecular dynamics and a united-residue (UNRES) model of polypeptide chains.J Mol Biol. 2006 Jan 20;355(3):536-47. doi: 10.1016/j.jmb.2005.10.056. Epub 2005 Nov 10. J Mol Biol. 2006. PMID: 16324712
-
Investigation of protein folding by coarse-grained molecular dynamics with the UNRES force field.J Phys Chem A. 2010 Apr 8;114(13):4471-85. doi: 10.1021/jp9117776. J Phys Chem A. 2010. PMID: 20166738 Free PMC article.
-
Folding processes of the B domain of protein A to the native state observed in all-atom ab initio folding simulations.J Chem Phys. 2008 Jun 21;128(23):235105. doi: 10.1063/1.2937135. J Chem Phys. 2008. PMID: 18570534 Free PMC article.
-
Modification and optimization of the united-residue (UNRES) potential energy function for canonical simulations. I. Temperature dependence of the effective energy function and tests of the optimization method with single training proteins.J Phys Chem B. 2007 Jan 11;111(1):260-85. doi: 10.1021/jp065380a. J Phys Chem B. 2007. PMID: 17201450 Free PMC article.
-
Folding dynamics of the Trp-cage miniprotein: evidence for a native-like intermediate from combined time-resolved vibrational spectroscopy and molecular dynamics simulations.J Phys Chem B. 2013 Oct 3;117(39):11490-501. doi: 10.1021/jp404714c. Epub 2013 Sep 19. J Phys Chem B. 2013. PMID: 24050152
Cited by
-
Exploring one-state downhill protein folding in single molecules.Proc Natl Acad Sci U S A. 2012 Jan 3;109(1):179-84. doi: 10.1073/pnas.1111164109. Epub 2011 Dec 19. Proc Natl Acad Sci U S A. 2012. PMID: 22184219 Free PMC article.
-
Flexible Target Recognition of the Intrinsically Disordered DNA-Binding Domain of CytR Monitored by Single-Molecule Fluorescence Spectroscopy.J Phys Chem B. 2022 Aug 25;126(33):6136-6147. doi: 10.1021/acs.jpcb.2c02791. Epub 2022 Aug 15. J Phys Chem B. 2022. PMID: 35969476 Free PMC article.
-
Coarse-grained force field: general folding theory.Phys Chem Chem Phys. 2011 Oct 14;13(38):16890-901. doi: 10.1039/c1cp20752k. Epub 2011 Jun 3. Phys Chem Chem Phys. 2011. PMID: 21643583 Free PMC article. Review.
-
Simulation of chaperonin effect on protein folding: a shift from nucleation-condensation to framework mechanism.J Am Chem Soc. 2011 Jul 6;133(26):10283-9. doi: 10.1021/ja203275f. Epub 2011 Jun 10. J Am Chem Soc. 2011. PMID: 21618995 Free PMC article.
-
Accounting for a mirror-image conformation as a subtle effect in protein folding.Proc Natl Acad Sci U S A. 2014 Jun 10;111(23):8458-63. doi: 10.1073/pnas.1407837111. Epub 2014 May 27. Proc Natl Acad Sci U S A. 2014. PMID: 24912167 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

