Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids
- PMID: 20569222
- PMCID: PMC3121328
- DOI: 10.2144/000113418
Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids
Abstract
Here we describe a straightforward, efficient, and reliable way to clone an insert of choice into a plasmid of choice without restriction endonucleases or T4 DNA ligase. Chimeric primers containing plasmid sequence at the 5' ends and insert sequence at the 3' ends were used to PCR-amplify insertion sequences of various sizes, namely the genes for GFP (gfp), beta-d-glucuronidase (gusA), and beta-galactosidase (lacZ), as well as the entire luxABCDE operon. These inserts were employed as mega-primers in a second PCR with a circular plasmid template. The original plasmid templates were then destroyed in restriction digests with DpnI, and the overlap extension PCR products were used to transform competent Escherichia coli cells. Phusion DNA polymerase was used for the amplification and fusion reactions, so both reactions were easy to monitor and optimize.
Conflict of interest statement
The authors declare no competing interests.
Figures
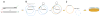
Comment in
-
Methods in 2012, part 2.Biotechniques. 2012 Jul;53(1):11. doi: 10.2144/000113882. Biotechniques. 2012. PMID: 22780309 No abstract available.
References
-
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. CSH Laboratory Press; Cold Spring Harbor, NY: 2001.
-
- Rashtchian A, Thornton CG, Heidecker G. A novel method for site-directed mutagenesis using PCR and uracil DNA glycosylase. PCR Methods Appl. 1992;2:124–130. - PubMed
-
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
