Everything you wanted to know about Markov State Models but were afraid to ask
- PMID: 20570730
- PMCID: PMC2933958
- DOI: 10.1016/j.ymeth.2010.06.002
Everything you wanted to know about Markov State Models but were afraid to ask
Abstract
Simulating protein folding has been a challenging problem for decades due to the long timescales involved (compared with what is possible to simulate) and the challenges of gaining insight from the complex nature of the resulting simulation data. Markov State Models (MSMs) present a means to tackle both of these challenges, yielding simulations on experimentally relevant timescales, statistical significance, and coarse grained representations that are readily humanly understandable. Here, we review this method with the intended audience of non-experts, in order to introduce the method to a broader audience. We review the motivations, methods, and caveats of MSMs, as well as some recent highlights of applications of the method. We conclude by discussing how this approach is part of a paradigm shift in how one uses simulations, away from anecdotal single-trajectory approaches to a more comprehensive statistical approach.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures
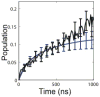
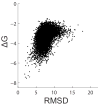
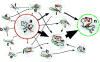
References
-
- Snow CD, et al. How well can simulation predict protein folding kinetics and thermodynamics? Annu Rev Biophys Biomol Struct. 2005;34:43–69. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

