Genomic and biological characterization of exon 4 KRAS mutations in human cancer
- PMID: 20570890
- PMCID: PMC2943514
- DOI: 10.1158/0008-5472.CAN-10-0192
Genomic and biological characterization of exon 4 KRAS mutations in human cancer
Abstract
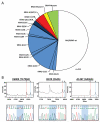
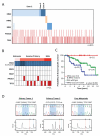
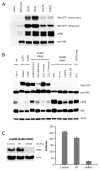
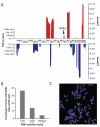
Mutations in RAS proteins occur widely in human cancer. Prompted by the confirmation of KRAS mutation as a predictive biomarker of response to epidermal growth factor receptor (EGFR)-targeted therapies, limited clinical testing for RAS pathway mutations has recently been adopted. We performed a multiplatform genomic analysis to characterize, in a nonbiased manner, the biological, biochemical, and prognostic significance of Ras pathway alterations in colorectal tumors and other solid tumor malignancies. Mutations in exon 4 of KRAS were found to occur commonly and to predict for a more favorable clinical outcome in patients with colorectal cancer. Exon 4 KRAS mutations, all of which were identified at amino acid residues K117 and A146, were associated with lower levels of GTP-bound RAS in isogenic models. These same mutations were also often accompanied by conversion to homozygosity and increased gene copy number, in human tumors and tumor cell lines. Models harboring exon 4 KRAS mutations exhibited mitogen-activated protein/extracellular signal-regulated kinase kinase dependence and resistance to EGFR-targeted agents. Our findings suggest that RAS mutation is not a binary variable in tumors, and that the diversity in mutant alleles and variability in gene copy number may also contribute to the heterogeneity of clinical outcomes observed in cancer patients. These results also provide a rationale for broader KRAS testing beyond the most common hotspot alleles in exons 2 and 3.
(c)2010 AACR.
Figures

References
-
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–65. - PubMed
-
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. - PubMed
-
- Chang EH, Furth ME, Scolnick EM, Lowy DR. Tumorigenic transformation of mammalian cells induced by a normal human gene homologous to the oncogene of Harvey murine sarcoma virus. Nature. 1982;297:479–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

