Modeling bistable cell-fate choices in the Drosophila eye: qualitative and quantitative perspectives
- PMID: 20570936
- PMCID: PMC2889600
- DOI: 10.1242/dev.044826
Modeling bistable cell-fate choices in the Drosophila eye: qualitative and quantitative perspectives
Abstract
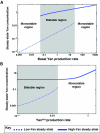
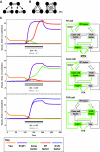
A major goal of developmental biology is to understand the molecular mechanisms whereby genetic signaling networks establish and maintain distinct cell types within multicellular organisms. Here, we review cell-fate decisions in the developing eye of Drosophila melanogaster and the experimental results that have revealed the topology of the underlying signaling circuitries. We then propose that switch-like network motifs based on positive feedback play a central role in cell-fate choice, and discuss how mathematical modeling can be used to understand and predict the bistable or multistable behavior of such networks.
Figures





References
-
- Amonlirdviman K., Khare N. A., Tree D. R. P., Chen W.-S., Axelrod J. D., Tomlin C. J. (2005). Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science 307, 423-426 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

