Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins
- PMID: 20571077
- PMCID: PMC2939488
- DOI: 10.1124/mol.110.065128
Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins
Abstract
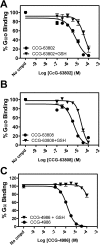
Regulators of G protein signaling (RGS) proteins are potent negative modulators of G protein signaling and have been proposed as potential targets for small-molecule inhibitor development. We report a high-throughput time-resolved fluorescence resonance energy transfer screen to identify inhibitors of RGS4 and describe the first reversible small-molecule inhibitors of an RGS protein. Two closely related compounds, typified by CCG-63802 [((2E)-2-(1,3-benzothiazol-2-yl)-3-[9-methyl-2-(3-methylphenoxy)-4-oxo-4H-pyrido[1,2-a]pyrimidin-3-yl]prop-2-enenitrile)], inhibit the interaction between RGS4 and Galpha(o) with an IC(50) value in the low micromolar range. They show selectivity among RGS proteins with a potency order of RGS 4 > 19 = 16 > 8 >> 7. The compounds inhibit the GTPase accelerating protein activity of RGS4, and thermal stability studies demonstrate binding to the RGS but not to Galpha(o). On RGS4, they depend on an interaction with one or more cysteines in a pocket that has previously been identified as an allosteric site for RGS regulation by acidic phospholipids. Unlike previous small-molecule RGS inhibitors identified to date, these compounds retain substantial activity under reducing conditions and are fully reversible on the 10-min time scale. CCG-63802 and related analogs represent a useful step toward the development of chemical tools for the study of RGS physiology.
Figures





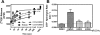
References
-
- Abad MC, Askari H, O'Neill J, Klinger AL, Milligan C, Lewandowski F, Springer B, Spurlino J, Rentzeperis D. (2008) Structural determination of estrogen-related receptor γ in the presence of phenol derivative compounds. J Steroid Biochem Mol Biol 108:44–54 - PubMed
-
- Arkin MR, Wells JA. (2004) Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 3:301–317 - PubMed
-
- Arkin MR, Whitty A. (2009) The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol 13:284–290 - PubMed
-
- Berg T. (2003) Modulation of protein-protein interactions with small organic molecules. Angew Chem Int Ed Engl 42:2462–2481 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

