Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA
- PMID: 20571084
- PMCID: PMC2965240
- DOI: 10.1093/nar/gkq521
Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA
Abstract
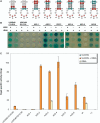
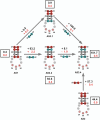
While a number of aminoacyl tRNA synthetase (aaRS):tRNA pairs have been engineered to alter or expand the genetic code, only the Methanococcus jannaschii tyrosyl tRNA synthetase and tRNA have been used extensively in bacteria, limiting the types and numbers of unnatural amino acids that can be utilized at any one time to expand the genetic code. In order to expand the number and type of aaRS/tRNA pairs available for engineering bacterial genetic codes, we have developed an orthogonal tryptophanyl tRNA synthetase and tRNA pair, derived from Saccharomyces cerevisiae. In the process of developing an amber suppressor tRNA, we discovered that the Escherichia coli lysyl tRNA synthetase was responsible for misacylating the initial amber suppressor version of the yeast tryptophanyl tRNA. It was discovered that modification of the G:C content of the anticodon stem and therefore reducing the structural flexibility of this stem eliminated misacylation by the E. coli lysyl tRNA synthetase, and led to the development of a functional, orthogonal suppressor pair that should prove useful for the incorporation of bulky, unnatural amino acids into the genetic code. Our results provide insight into the role of tRNA flexibility in molecular recognition and the engineering and evolution of tRNA specificity.
Figures



References
-
- Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Nakayama H, Takio K, et al. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl Acad. Sci. USA. 2002;99:9715–9720. - PMC - PubMed
-
- Xie J, Schultz PG. A chemical toolkit for proteins–an expanded genetic code. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. - PubMed
-
- Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. - PubMed
-
- Pastrnak M, Mageliery TM, Schultz PG. A new orthogonal suppressor tRNA/Aminoacyl-tRNA synthetase pair for evolving an organism with an expanded genetic code. Helv. Chim. Acta. 2000;83:2277–2286.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

