In silico structure-function analysis of pathological variation in the HSD11B2 gene sequence
- PMID: 20571110
- PMCID: PMC2929884
- DOI: 10.1152/physiolgenomics.00053.2010
In silico structure-function analysis of pathological variation in the HSD11B2 gene sequence
Abstract
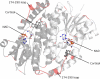
11beta-Hydroxysteroid dehydrogenase type 2 (11betaHSD2) is a short-chain dehydrogenase/reductase (SDR) responsible for inactivating cortisol and preventing its binding to the mineralocorticoid receptor (MR). Nonfunctional mutations in HSD11B2, the gene encoding 11betaHSD2, cause the hypertensive syndrome of apparent mineralocorticoid excess (AME). Like other such Mendelian disorders, AME is rare but has nevertheless helped to illuminate principles fundamental to the regulation of blood pressure. Furthermore, polymorphisms in HSD11B2 have been associated with salt sensitivity, a major risk factor for cardiovascular mortality. It is therefore highly likely that sequence variation in HSD11B2, having subtle functional ramifications, will affect blood pressure in the wider population. In this study, a three-dimensional homology model of 11betaHSD2 was created and used to hypothesize the functional consequences in terms of protein structure of published mutations in HSD11B2. This approach underscored the strong genotype-phenotype correlation of AME: severe forms of the disease, associated with little in vivo enzyme activity, arise from mutations occurring in invariant alignment positions. These were predicted to exert gross structural changes in the protein. In contrast, those mutations causing a mild clinical phenotype were in less conserved regions of the protein that were predicted to be relatively more tolerant to substitution. Finally, a number of pathogenic mutations are shown to be associated with regions predicted to participate in dimer formation, and in protein stabilization, which may therefore suggest molecular mechanisms of disease.
Figures








Similar articles
-
A novel mutation in HSD11B2 causes apparent mineralocorticoid excess in an Omani kindred.Ann N Y Acad Sci. 2016 Jul;1376(1):65-71. doi: 10.1111/nyas.13162. Epub 2016 Aug 15. Ann N Y Acad Sci. 2016. PMID: 27526338
-
Conditional Deletion of Hsd11b2 in the Brain Causes Salt Appetite and Hypertension.Circulation. 2016 Apr 5;133(14):1360-70. doi: 10.1161/CIRCULATIONAHA.115.019341. Epub 2016 Mar 7. Circulation. 2016. PMID: 26951843 Free PMC article.
-
Apparent Mineralocorticoid Excess by a Novel Mutation and Epigenetic Modulation by HSD11B2 Promoter Methylation.J Clin Endocrinol Metab. 2015 Sep;100(9):E1234-41. doi: 10.1210/jc.2015-1760. Epub 2015 Jun 30. J Clin Endocrinol Metab. 2015. PMID: 26126204
-
Apparent mineralocorticoid excess caused by novel compound heterozygous mutations in HSD11B2 and characterized by early-onset hypertension and hypokalemia.Endocrine. 2020 Dec;70(3):607-615. doi: 10.1007/s12020-020-02460-9. Epub 2020 Aug 20. Endocrine. 2020. PMID: 32816205 Free PMC article.
-
The role of 11β-hydroxysteroid dehydrogenase type 2 in human hypertension.Biochim Biophys Acta. 2010 Dec;1802(12):1178-87. doi: 10.1016/j.bbadis.2009.10.017. Epub 2009 Nov 10. Biochim Biophys Acta. 2010. PMID: 19909806 Review.
Cited by
-
Structural comparison, substrate specificity, and inhibitor binding of AGPase small subunit from monocot and dicot: present insight and future potential.Biomed Res Int. 2014;2014:583606. doi: 10.1155/2014/583606. Epub 2014 Sep 2. Biomed Res Int. 2014. PMID: 25276800 Free PMC article.
-
11β-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action.Physiol Rev. 2013 Jul;93(3):1139-206. doi: 10.1152/physrev.00020.2012. Physiol Rev. 2013. PMID: 23899562 Free PMC article. Review.
-
Case report: Clinical characteristics and Genetical analysis of HSD11B2 in three Chinese children with apparent mineralocorticoid excess: a case series.Front Endocrinol (Lausanne). 2025 Jan 27;15:1491825. doi: 10.3389/fendo.2024.1491825. eCollection 2024. Front Endocrinol (Lausanne). 2025. PMID: 39931437 Free PMC article.
-
Apparent mineralocorticoid excess: comprehensive overview of molecular genetics.J Transl Med. 2022 Nov 3;20(1):500. doi: 10.1186/s12967-022-03698-9. J Transl Med. 2022. PMID: 36329487 Free PMC article. Review.
-
Structure-Function Relationships of LDL Receptor Missense Mutations Using Homology Modeling.Protein J. 2019 Aug;38(4):447-462. doi: 10.1007/s10930-019-09860-5. Protein J. 2019. PMID: 31401775
References
-
- Agarwal AK, Giacchetti G, Lavery G, Nikkila H, Palermo M, Ricketts M, McTernan C, Bianchi G, Manunta P, Strazzullo P, Mantero F, White PC, Stewart PM. CA-repeat polymorphism in intron 1 of HSD11B2: effects on gene expression and salt sensitivity. Hypertension 36: 187–194, 2000 - PubMed
-
- Alikhani-Koupaei R, Fouladkou F, Fustier P, Cenni B, Sharma AM, Deter H, Frey BM, Frey FJ. Identification of polymorphisms in the human 11beta-hydroxysteroid dehydrogenase type 2 gene promoter: functional characterization and relevance for salt sensitivity. FASEB J 21: 3618–3628, 2007 - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 215: 403–410, 1990 - PubMed
-
- Arnold P, Tam S, Yan L, Baker ME, Frey FJ, Odermatt A. Glutamate-115 renders specificity of human 11beta-hydroxysteroid dehydrogenase type 2 for the cofactor NAD+. Mol Cell Endocrinol 201: 177–187, 2003 - PubMed
-
- Arriza JL, Weinberger C, Cerelli G, Glaser TM, Handelin BL, Housman DE, Evans RM. Cloning of human mineralocorticoid receptor complementary DNA: structural and functional kinship with the glucocorticoid receptor. Science 237: 268–275, 1987 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

