ATP-sensitive K(+) channel-deficient dilated cardiomyopathy proteome remodeled by embryonic stem cell therapy
- PMID: 20572010
- PMCID: PMC3129996
- DOI: 10.1002/stem.465
ATP-sensitive K(+) channel-deficient dilated cardiomyopathy proteome remodeled by embryonic stem cell therapy
Abstract
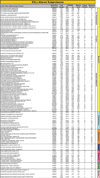
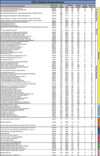
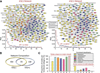
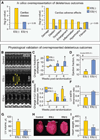
Transplantation of pluripotent stem cells has proven beneficial in heart failure, yet the proteomic landscape underlying repair remains largely uncharacterized. In a genetic model of dilated cardiomyopathy elicited by pressure overload in the KCNJ11 (potassium inwardly rectifying channel, subfamily J, member 11) null mutant, proteome-wide profiles were here resolved by means of a systems approach prior to and following disease manifestation in the absence or presence of embryonic stem cell treatment. Comparative two-dimensional gel electrophoresis revealed a unique cardiomyopathic proteome in the absence of therapy, remodeled in response to stem cell treatment. Specifically, linear ion trap quadrupole-Orbitrap mass spectrometry determined the identities of 93 and 109 differentially expressed proteins from treated and untreated cardiomyopathic hearts, respectively. Mapped protein-protein relationships and corresponding neighborhoods incorporated the stem cell-dependent subproteome into a nonstochastic network with divergent composition from the stem cell-independent counterpart. Stem cell intervention produced a distinct proteome signature across a spectrum of biological processes ranging from energetic metabolism, oxidoreductases, and stress-related chaperones to processes supporting protein synthesis/degradation, signaling, and transport regulation, cell structure and scaffolding. In the absence of treatment, bioinformatic interrogation of the disease-only proteome network prioritized adverse cardiac outcomes, ablated or ameliorated following stem cell transplantation. Functional and structural measurements validated improved myocardial contractile performance, reduced ventricular size and decreased cardiac damage in the treated cohort. Unbiased systems assessment unmasked "cardiovascular development" as a prioritized biological function in stem cell-reconstructed cardiomyopathic hearts. Thus, embryonic stem cell treatment transformed the cardiomyopathic proteome to demote disease-associated adverse effects and sustain a procardiogenic developmental response, supplying a regenerative substrate for heart failure repair.
Conflict of interest statement
The authors indicate no potential conflicts of interest.
Figures


Similar articles
-
Proteomic profiling of KATP channel-deficient hypertensive heart maps risk for maladaptive cardiomyopathic outcome.Proteomics. 2009 Mar;9(5):1314-25. doi: 10.1002/pmic.200800718. Proteomics. 2009. PMID: 19253285 Free PMC article.
-
ATP-sensitive K+ channel knockout induces cardiac proteome remodeling predictive of heart disease susceptibility.J Proteome Res. 2009 Oct;8(10):4823-34. doi: 10.1021/pr900561g. J Proteome Res. 2009. PMID: 19673485 Free PMC article.
-
K(ATP) channel-dependent metaboproteome decoded: systems approaches to heart failure prediction, diagnosis, and therapy.Cardiovasc Res. 2011 May 1;90(2):258-66. doi: 10.1093/cvr/cvr046. Epub 2011 Feb 14. Cardiovasc Res. 2011. PMID: 21321057 Free PMC article. Review.
-
Mechanical dyssynchrony precedes QRS widening in ATP-sensitive K⁺ channel-deficient dilated cardiomyopathy.J Am Heart Assoc. 2013 Dec 5;2(6):e000410. doi: 10.1161/JAHA.113.000410. J Am Heart Assoc. 2013. PMID: 24308936 Free PMC article.
-
Human K(ATP) channelopathies: diseases of metabolic homeostasis.Pflugers Arch. 2010 Jul;460(2):295-306. doi: 10.1007/s00424-009-0771-y. Epub 2009 Dec 24. Pflugers Arch. 2010. PMID: 20033705 Free PMC article. Review.
Cited by
-
Systems proteomics for translational network medicine.Circ Cardiovasc Genet. 2012 Aug 1;5(4):478. doi: 10.1161/CIRCGENETICS.110.958991. Circ Cardiovasc Genet. 2012. PMID: 22896016 Free PMC article.
-
Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming.Cell Metab. 2011 Aug 3;14(2):264-71. doi: 10.1016/j.cmet.2011.06.011. Cell Metab. 2011. PMID: 21803296 Free PMC article.
-
Metabolome and metaboproteome remodeling in nuclear reprogramming.Cell Cycle. 2013 Aug 1;12(15):2355-65. doi: 10.4161/cc.25509. Epub 2013 Jul 8. Cell Cycle. 2013. PMID: 23839047 Free PMC article.
-
Stem cell systems informatics for advanced clinical biodiagnostics: tracing molecular signatures from bench to bedside.Croat Med J. 2013 Aug;54(4):319-29. doi: 10.3325//cmj.2013.54.319. Croat Med J. 2013. PMID: 23986272 Free PMC article. Review.
-
A mighty small heart: the cardiac proteome of adult Drosophila melanogaster.PLoS One. 2011 Apr 25;6(4):e18497. doi: 10.1371/journal.pone.0018497. PLoS One. 2011. PMID: 21541028 Free PMC article.
References
-
- Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. - PubMed
-
- Hershberger RE, Lindenfeld J, Mestroni L, et al. Genetic evaluation of cardiomyopathy: A Heart Failure Society of America practice guideline. J Card Fail. 2009;15:83–97. - PubMed
-
- Luk A, Ahn E, Soor GS, et al. Dilated cardiomyopathy: A review. J Clin Pathol. 2009;62:219–225. - PubMed
-
- Chien KR. Genomic circuits and the integrative biology of cardiac diseases. Nature. 2000;407:227–232. - PubMed
-
- Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

