NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
- PMID: 20572013
- PMCID: PMC2923510
- DOI: 10.1002/pro.438
NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Abstract
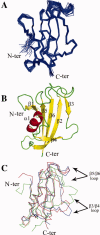
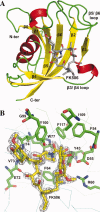
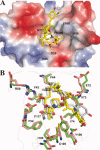
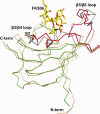
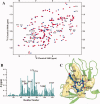
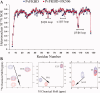
The emergence of drug-resistant malaria parasites is the major threat to effective malaria control, prompting a search for novel compounds with mechanisms of action that are different from the traditionally used drugs. The immunosuppressive drug FK506 shows an antimalarial activity. The mechanism of the drug action involves the molecular interaction with the parasite target proteins PfFKBP35 and PvFKBP35, which are novel FK506 binding protein family (FKBP) members from Plasmodium falciparum and Plasmodium vivax, respectively. Currently, molecular mechanisms of the FKBP family proteins in the parasites still remain elusive. To understand their functions, here we have determined the structures of the FK506 binding domain of Plasmodium vivax (PvFKBD) in unliganded form by NMR spectroscopy and in complex with FK506 by X-ray crystallography. We found out that PvFKBP35 exhibits a canonical FKBD fold and shares kinetic profiles similar to those of PfFKBP35, the homologous protein in P. falciparum, indicating that the parasite FKBP family members play similar biological roles in their life cycles. Despite the similarity, differences were observed in the ligand binding modes between PvFKBD and HsFKBP12, a human FKBP homolog, which could provide insightful information into designing selective antimalarial drug against the parasites.
Figures
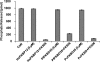
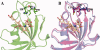
Similar articles
-
Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax.Eukaryot Cell. 2013 Apr;12(4):627-34. doi: 10.1128/EC.00016-13. Epub 2013 Feb 22. Eukaryot Cell. 2013. PMID: 23435727 Free PMC article.
-
Crystal structure of the FK506 binding domain of Plasmodium falciparum FKBP35 in complex with FK506.Biochemistry. 2008 Jun 3;47(22):5951-61. doi: 10.1021/bi800004u. Epub 2008 May 9. Biochemistry. 2008. PMID: 18465874
-
NMR assignments of the FK506-binding domain of FK506-binding protein 35 from Plasmodium vivax.Biomol NMR Assign. 2009 Dec;3(2):243-5. doi: 10.1007/s12104-009-9185-1. Biomol NMR Assign. 2009. PMID: 19774494
-
Structural insights into Plasmodium PPIases.Front Cell Infect Microbiol. 2022 Sep 2;12:931635. doi: 10.3389/fcimb.2022.931635. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36118020 Free PMC article. Review.
-
FKBP family proteins: immunophilins with versatile biological functions.Neurosignals. 2008;16(4):318-25. doi: 10.1159/000123041. Epub 2008 Jul 18. Neurosignals. 2008. PMID: 18635947 Review.
Cited by
-
Structural insights into substrate binding by PvFKBP35, a peptidylprolyl cis-trans isomerase from the human malarial parasite Plasmodium vivax.Eukaryot Cell. 2013 Apr;12(4):627-34. doi: 10.1128/EC.00016-13. Epub 2013 Feb 22. Eukaryot Cell. 2013. PMID: 23435727 Free PMC article.
-
Genetic validation of PfFKBP35 as an antimalarial drug target.Elife. 2023 Nov 7;12:RP86975. doi: 10.7554/eLife.86975. Elife. 2023. PMID: 37934560 Free PMC article.
-
High-resolution crystal structure of FKBP12 from Aedes aegypti.Protein Sci. 2012 Jul;21(7):1080-4. doi: 10.1002/pro.2079. Epub 2012 May 18. Protein Sci. 2012. PMID: 22517662 Free PMC article.
-
Small molecule Plasmodium FKBP35 inhibitor as a potential antimalaria agent.Sci Rep. 2013;3:2501. doi: 10.1038/srep02501. Sci Rep. 2013. PMID: 23974147 Free PMC article.
-
A systematic and prospectively validated approach for identifying synergistic drug combinations against malaria.Malar J. 2018 Apr 11;17(1):160. doi: 10.1186/s12936-018-2294-5. Malar J. 2018. PMID: 29642892 Free PMC article.
References
-
- World Health Organization (2005) World Malaria Report.
-
- Pain A, Hertz-Fowler C. Plasmodium genomics: latest milestone. Nat Rev Microbiol. 2009;7:180–181. - PubMed
-
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, Cheng Q, Coulson RM, Crabb BS, Del Portillo HA, Essien K, Feldblyum TV, Fernandez-Becerra C, Gilson PR, Gueye AH, Guo X, Kang'a S, Kooij TW, Korsinczky M, Meyer EV, Nene V, Paulsen I, White O, Ralph SA, Ren Q, Sargeant TJ, Salzberg SL, Stoeckert CJ, Sullivan SA, Yamamoto MM, Hoffman SL, Wortman JR, Gardner MJ, Galinski MR, Barnwell JW, Fraser-Liggett CM. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757–763. - PMC - PubMed
-
- Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, Kochar A, Khatri MP, Gupta V. Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am J Trop Med Hyg. 2009;80:194–198. - PubMed
-
- Hyde JE. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 2002;4:165–174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

