Topology of the disulfide bonds in the antiviral lectin scytovirin
- PMID: 20572021
- PMCID: PMC2975129
- DOI: 10.1002/pro.445
Topology of the disulfide bonds in the antiviral lectin scytovirin
Abstract
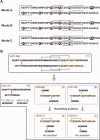
The antiviral lectin scytovirin (SVN) contains a total of five disulfide bonds in two structurally similar domains. Previous reports provided contradictory results on the disulfide pairing in each individual domain, and we have now re-examined the disulfide topology. N-terminal sequencing and mass spectrometry were used to analyze proteolytic fragments of native SVN obtained at acidic pH, yielding the assignment as Cys7-Cys55, Cys20-Cys32, Cys26-Cys38, Cys68-Cys80, and Cys74-Cys86. We also analyzed the N-terminal domain of SVN (SD1, residues 1-48) prepared by expression/oxidative folding of the recombinant protein and by chemical synthesis. The disulfide pairing in the chemically synthesized SD1 was forced into predetermined topologies: SD1A (Cys20-Cys26, Cys32-Cys38) or SD1B (Cys20-Cys32, Cys26-Cys38). The topology of native SVN was found to be in agreement with the SD1B and the one determined for the recombinant SD1 domain. Although the two synthetic forms of SD1 were distinct when subjected to chromatography, their antiviral properties were indistinguishable, having low nM activity against HIV. Tryptic fragments, the "cystine clusters" [Cys20-Cys32/Cys26-Cys38; SD1] and [Cys68-Cys80/Cys74-C-86; SD2], were found to undergo rapid disulfide interchange at pH 8. This interchange resulted in accumulation of artifactual fragments in alkaline pH digests that are structurally unrelated to the original topology, providing a rational explanation for the differences between the topology reported herein and the one reported earlier (Bokesh et al., Biochemistry 2003;42:2578-2584). Our observations emphasize the fact that proteins such as SVN, with disulfide bonds in close proximity, require considerable precautions when being fragmented for the purpose of disulfide assignment.
Copyright © 2010 The Protein Society.
Figures





Similar articles
-
In silico analysis of the cyanobacterial lectin scytovirin: new insights into binding properties.Mol Biol Rep. 2017 Aug;44(4):353-358. doi: 10.1007/s11033-017-4116-1. Epub 2017 Jul 29. Mol Biol Rep. 2017. PMID: 28756560 Free PMC article.
-
Antiviral Cyanometabolites-A Review.Biomolecules. 2021 Mar 22;11(3):474. doi: 10.3390/biom11030474. Biomolecules. 2021. PMID: 33810129 Free PMC article. Review.
-
Potent anti-HIV activity of scytovirin domain 1 peptide.Peptides. 2006 Jul;27(7):1668-75. doi: 10.1016/j.peptides.2006.03.018. Epub 2006 May 2. Peptides. 2006. PMID: 16647158
-
The novel fold of scytovirin reveals a new twist for antiviral entry inhibitors.J Mol Biol. 2007 Jun 1;369(2):451-61. doi: 10.1016/j.jmb.2007.03.030. Epub 2007 Mar 20. J Mol Biol. 2007. PMID: 17434526 Free PMC article.
-
Cyanobacterial lectins characteristics and their role as antiviral agents.Int J Biol Macromol. 2017 Sep;102:475-496. doi: 10.1016/j.ijbiomac.2017.04.041. Epub 2017 Apr 24. Int J Biol Macromol. 2017. PMID: 28437766 Review.
Cited by
-
In silico analysis of the cyanobacterial lectin scytovirin: new insights into binding properties.Mol Biol Rep. 2017 Aug;44(4):353-358. doi: 10.1007/s11033-017-4116-1. Epub 2017 Jul 29. Mol Biol Rep. 2017. PMID: 28756560 Free PMC article.
-
Recent mass spectrometry-based techniques and considerations for disulfide bond characterization in proteins.Anal Bioanal Chem. 2018 Apr;410(10):2467-2484. doi: 10.1007/s00216-017-0772-1. Epub 2017 Dec 18. Anal Bioanal Chem. 2018. PMID: 29256076 Free PMC article. Review.
-
Antiviral Cyanometabolites-A Review.Biomolecules. 2021 Mar 22;11(3):474. doi: 10.3390/biom11030474. Biomolecules. 2021. PMID: 33810129 Free PMC article. Review.
-
HIV-1 enhancing effect of prostatic acid phosphatase peptides is reduced in human seminal plasma.PLoS One. 2011 Jan 20;6(1):e16285. doi: 10.1371/journal.pone.0016285. PLoS One. 2011. PMID: 21283773 Free PMC article.
-
Lectins and lectibodies: potential promising antiviral agents.Cell Mol Biol Lett. 2022 May 13;27(1):37. doi: 10.1186/s11658-022-00338-4. Cell Mol Biol Lett. 2022. PMID: 35562647 Free PMC article. Review.
References
-
- Bokesch HR, O'Keefe BR, McKee TC, Pannell LK, Patterson GM, Gardella RS, Sowder RC, Turpin J, Watson K, Buckheit RW, Jr, Boyd MR. A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Biochemistry. 2003;42:2578–2584. - PubMed
-
- Xiong C, O'Keefe BR, Byrd RA, McMahon JB. Potent anti-HIV activity of scytovirin domain 1 peptide. Peptides. 2006;27:1668–1675. - PubMed
-
- Xiong C, O'Keefe BR, Botos I, Wlodawer A, McMahon JB. Overexpression and purification of scytovirin, a potent, novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Protein Expr Purif. 2006;46:233–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

