Escape from X inactivation in mice and humans
- PMID: 20573260
- PMCID: PMC2911101
- DOI: 10.1186/gb-2010-11-6-213
Escape from X inactivation in mice and humans
Abstract
A subset of X-linked genes escapes silencing by X inactivation and is expressed from both X chromosomes in mammalian females. Species-specific differences in the identity of these genes have recently been discovered, suggesting a role in the evolution of sex differences. Chromatin analyses have aimed to discover how genes remain expressed within a repressive environment.
Figures

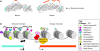
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

