Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment
- PMID: 20573702
- PMCID: PMC2927698
- DOI: 10.1242/dev.048363
Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment
Abstract
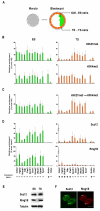
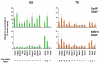
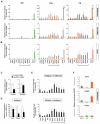
Pluripotent cells develop within the inner cell mass of blastocysts, a mosaic of cells surrounded by an extra-embryonic layer, the trophectoderm. We show that a set of somatic lineage regulators (including Hox, Gata and Sox factors) that carry bivalent chromatin enriched in H3K27me3 and H3K4me2 are selectively targeted by Suv39h1-mediated H3K9me3 and de novo DNA methylation in extra-embryonic versus embryonic (pluripotent) lineages, as assessed both in blastocyst-derived stem cells and in vivo. This stably repressed state is linked with a loss of gene priming for transcription through the exclusion of PRC1 (Ring1B) and RNA polymerase II complexes at bivalent, lineage-inappropriate genes upon trophoblast lineage commitment. Collectively, our results suggest a mutually exclusive role for Ring1B and Suv39h1 in regulating distinct chromatin states at key developmental genes and propose a novel mechanism by which lineage specification can be reinforced during early development.
Figures


References
-
- Arney K. L., Bao S., Bannister A. J., Kouzarides T., Surani M. A. (2002). Histone methylation defines epigenetic asymmetry in the mouse zygote. Int. J. Dev. Biol. 46, 317-320 - PubMed
-
- Atsuta T., Fujimura S., Moriya H., Vidal M., Akasaka T., Koseki H. (2001). Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma 20, 43-46 - PubMed
-
- Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M., et al. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532-538 - PubMed
-
- Bantignies F., Cavalli G. (2006). Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 18, 275-283 - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823-837 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

