Paraspeckles
- PMID: 20573717
- PMCID: PMC2890200
- DOI: 10.1101/cshperspect.a000687
Paraspeckles
Abstract
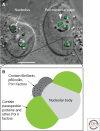
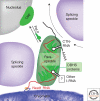
Paraspeckles are a relatively new class of subnuclear bodies found in the interchromatin space of mammalian cells. They are RNA-protein structures formed by the interaction between a long nonprotein-coding RNA species, NEAT1/Men epsilon/beta, and members of the DBHS (Drosophila Behavior Human Splicing) family of proteins: P54NRB/NONO, PSPC1, and PSF/SFPQ. Paraspeckles are critical to the control of gene expression through the nuclear retention of RNA containing double-stranded RNA regions that have been subject to adenosine-to-inosine editing. Through this mechanism paraspeckles and their components may ultimately have a role in controlling gene expression during many cellular processes including differentiation, viral infection, and stress responses.
Figures


References
-
- Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen H, Mann M, Lamond AI 2002. Directed proteomic analysis of the human nucleolus. Current Biology 12:1–11 - PubMed
-
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, Schibler U 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308:693–696 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
