The triplet repeats of the Sin Nombre hantavirus 5' untranslated region are sufficient in cis for nucleocapsid-mediated translation initiation
- PMID: 20573811
- PMCID: PMC2918995
- DOI: 10.1128/JVI.02720-09
The triplet repeats of the Sin Nombre hantavirus 5' untranslated region are sufficient in cis for nucleocapsid-mediated translation initiation
Abstract
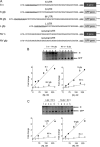
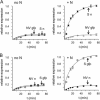
Hantavirus nucleocapsid protein (N) can replace the cellular cap-binding complex, eukaryotic initiation factor 4F (eIF4F), to mediate translation initiation. Although N can augment translation initiation of nonviral mRNA, initiation of viral mRNA by N is superior. All members of the Bunyaviridae family, including the species of the hantavirus genus, express either three or four primary mRNAs from their tripartite negative-sense genomes. The 5' ends of the mRNAs contain nonviral heterologous oligonucleotides that originate from endonucleolytic cleavage of cellular mRNA during the process of cap snatching. In the hantaviruses these caps terminate with a 3' G residue followed by nucleotides arising from the viral template. Further, the 5' untranslated region (UTR) of viral mRNA uniformly contains, near the 5' end, either two or three copies of the triplet repeat sequence, UAGUAG or UAGUAGUAG. Through analysis of a panel of mutants with mutations in the viral UTR, we found that the sequence GUAGUAG is sufficient for preferential N-mediated translation initiation and for high-affinity binding of N to the UTR. This heptanucleotide sequence is present in viral mRNA containing either two or three copies of the triplet repeat.
Figures



Similar articles
-
Modulations in the host cell proteome by the hantavirus nucleocapsid protein.PLoS Pathog. 2024 Jan 8;20(1):e1011925. doi: 10.1371/journal.ppat.1011925. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38190410 Free PMC article.
-
Signatures of host mRNA 5' terminus for efficient hantavirus cap snatching.J Virol. 2012 Sep;86(18):10173-85. doi: 10.1128/JVI.05560-11. Epub 2012 Jul 11. J Virol. 2012. PMID: 22787213 Free PMC article.
-
Characterization of the Interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19).J Biol Chem. 2011 Apr 1;286(13):11814-24. doi: 10.1074/jbc.M110.210179. Epub 2011 Feb 4. J Biol Chem. 2011. PMID: 21296889 Free PMC article.
-
An atypical IRES within the 5' UTR of a dicistrovirus genome.Virus Res. 2009 Feb;139(2):157-65. doi: 10.1016/j.virusres.2008.07.017. Epub 2008 Sep 11. Virus Res. 2009. PMID: 18755228 Review.
-
Serological diagnosis with recombinant N antigen for hantavirus infection.Virus Res. 2014 Jul 17;187:77-83. doi: 10.1016/j.virusres.2013.12.040. Epub 2014 Jan 31. Virus Res. 2014. PMID: 24487183 Review.
Cited by
-
Modulations in the host cell proteome by the hantavirus nucleocapsid protein.PLoS Pathog. 2024 Jan 8;20(1):e1011925. doi: 10.1371/journal.ppat.1011925. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38190410 Free PMC article.
-
Recent advances in hantavirus molecular biology and disease.Adv Appl Microbiol. 2011;74:35-75. doi: 10.1016/B978-0-12-387022-3.00006-9. Adv Appl Microbiol. 2011. PMID: 21459193 Free PMC article.
-
SARS-CoV-2 Nucleocapsid Protein Is a Potential Therapeutic Target for Anticoronavirus Drug Discovery.Microbiol Spectr. 2023 Jun 15;11(3):e0118623. doi: 10.1128/spectrum.01186-23. Epub 2023 May 18. Microbiol Spectr. 2023. PMID: 37199631 Free PMC article.
-
Orthohantaviruses: An Overview of the Current Status of Diagnostics and Surveillance.Viruses. 2025 Apr 26;17(5):622. doi: 10.3390/v17050622. Viruses. 2025. PMID: 40431633 Free PMC article. Review.
-
Crimean-Congo Hemorrhagic Fever Virus Nucleocapsid Protein Augments mRNA Translation.J Virol. 2017 Jul 12;91(15):e00636-17. doi: 10.1128/JVI.00636-17. Print 2017 Aug 1. J Virol. 2017. PMID: 28515298 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

