Visualizing the replication cycle of bunyamwera orthobunyavirus expressing fluorescent protein-tagged Gc glycoprotein
- PMID: 20573824
- PMCID: PMC2919021
- DOI: 10.1128/JVI.00902-10
Visualizing the replication cycle of bunyamwera orthobunyavirus expressing fluorescent protein-tagged Gc glycoprotein
Abstract
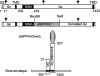
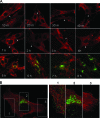
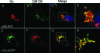
The virion glycoproteins Gn and Gc of Bunyamwera virus (BUNV), the prototype of the Bunyaviridae family and also of the Orthobunyavirus genus, are encoded by the medium (M) RNA genome segment and are involved in both viral attachment and entry. After their synthesis Gn and Gc form a heterodimer in the endoplasmic reticulum (ER) and transit to the Golgi compartment for virus assembly. The N-terminal half of the Gc ectodomain was previously shown to be dispensable for virus replication in cell culture (X. Shi, J. Goli, G. Clark, K. Brauburger, and R. M. Elliott, J. Gen. Virol. 90:2483-2492, 2009.). In this study, the coding sequence for a fluorescent protein, either enhanced green fluorescent protein (eGFP) or mCherry fluorescent protein, was fused to the N terminus of truncated Gc, and two recombinant BUNVs (rBUNGc-eGFP and rBUNGc-mCherry) were rescued by reverse genetics. The recombinant viruses showed bright autofluorescence under UV light and were competent for replication in various mammalian cell lines. rBUNGc-mCherry was completely stable over 10 passages, whereas internal, in-frame deletions occurred in the chimeric Gc-eGFP protein of rBUNGc-eGFP, resulting in loss of fluorescence between passages 5 and 7. Autofluorescence of the recombinant viruses allowed visualization of different stages of the infection cycle, including virus attachment to the cell surface, budding of virus particles in Golgi membranes, and virus-induced morphological changes to the Golgi compartment at later stages of infection. The fluorescent protein-tagged viruses will be valuable reagents for live-cell imaging studies to investigate virus entry, budding, and morphogenesis in real time.
Figures
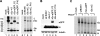

References
-
- Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

