Nucleocytoplasmic shuttling of human cytomegalovirus UL84 is essential for virus growth
- PMID: 20573826
- PMCID: PMC2919040
- DOI: 10.1128/JVI.00738-10
Nucleocytoplasmic shuttling of human cytomegalovirus UL84 is essential for virus growth
Abstract
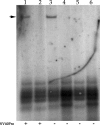
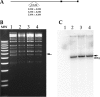
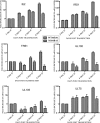
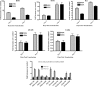
Human cytomegalovirus (HCMV) UL84 is a multifunctional protein that is the proposed initiator for lytic viral DNA synthesis. Recently it was shown that UL84 displays nucleocytoplasmic shuttling. The role of shuttling in lytic DNA replication and virus growth is unknown. We now show that expression of the nonshuttling UL84 mutant failed to complement oriLyt-dependent DNA replication in the transient assay under conditions where core replication and ancillary proteins were expressed under the control of their native promoters. However, constitutive expression of the core replication proteins, along with the nonshuttling UL84 mutant, resulted in efficient oriLyt amplification, suggesting that shuttling may contribute to the activity of one of the auxiliary replication proteins. A recombinant HCMV bacterial artificial chromosome plasmid (BACmid) expressing the nonshuttling UL84 mutant (NS84 BAC) was defective for production of infectious virus. Quantitative PCR showed that NS84 BAC had decreased accumulation of viral DNA in both cellular and supernatant samples. Analysis of the accumulation of select viral mRNAs showed no difference in total cellular mRNA accumulation for IE2, IRS1, TRS1, UL102, UL105, and UL75 in cells transfected with the NS84 BAC. However, examination of cytoplasmic RNA and subcellular localization of IRS1 revealed a decrease in IRS1 mRNA accumulation and displaced protein localization, strongly suggesting that UL84 facilitated the localization of IRS1 mRNA to the cytoplasm. RNA pulldown assays showed that UL84 interacted with IRS1 mRNA. These results indicate that nucleocytoplasmic shuttling is essential for virus growth and strongly suggest that UL84 is responsible for localization of at least one virus-encoded transcript, IRS1 mRNA.
Figures



Similar articles
-
Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth.J Virol. 2004 Oct;78(19):10360-9. doi: 10.1128/JVI.78.19.10360-10369.2004. J Virol. 2004. PMID: 15367602 Free PMC article.
-
Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays.J Virol. 1996 Nov;70(11):7398-413. doi: 10.1128/JVI.70.11.7398-7413.1996. J Virol. 1996. PMID: 8892858 Free PMC article.
-
Analysis of the interactions of viral and cellular factors with human cytomegalovirus lytic origin of replication, oriLyt.Virology. 2012 Mar 15;424(2):106-14. doi: 10.1016/j.virol.2011.12.010. Epub 2012 Jan 10. Virology. 2012. PMID: 22236369 Free PMC article.
-
Nuts and bolts of human cytomegalovirus lytic DNA replication.Curr Top Microbiol Immunol. 2008;325:153-66. doi: 10.1007/978-3-540-77349-8_9. Curr Top Microbiol Immunol. 2008. PMID: 18637505 Review.
-
Regulation of nucleocytoplasmic trafficking of viral proteins: an integral role in pathogenesis?Biochim Biophys Acta. 2011 Dec;1813(12):2176-90. doi: 10.1016/j.bbamcr.2011.03.019. Epub 2011 Apr 16. Biochim Biophys Acta. 2011. PMID: 21530593 Free PMC article. Review.
Cited by
-
The functions of herpesvirus shuttling proteins in the virus lifecycle.Front Microbiol. 2025 Feb 5;16:1515241. doi: 10.3389/fmicb.2025.1515241. eCollection 2025. Front Microbiol. 2025. PMID: 39973925 Free PMC article. Review.
-
Functional annotation of human cytomegalovirus gene products: an update.Front Microbiol. 2014 May 19;5:218. doi: 10.3389/fmicb.2014.00218. eCollection 2014. Front Microbiol. 2014. PMID: 24904534 Free PMC article. Review.
-
Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells.PLoS Pathog. 2013;9(5):e1003366. doi: 10.1371/journal.ppat.1003366. Epub 2013 May 23. PLoS Pathog. 2013. PMID: 23717203 Free PMC article. Clinical Trial.
-
Strategies for the Viral Exploitation of Nuclear Pore Transport Pathways.Viruses. 2025 Jan 23;17(2):151. doi: 10.3390/v17020151. Viruses. 2025. PMID: 40006906 Free PMC article. Review.
-
How SARS-CoV-2 and Other Viruses Build an Invasion Route to Hijack the Host Nucleocytoplasmic Trafficking System.Cells. 2021 Jun 7;10(6):1424. doi: 10.3390/cells10061424. Cells. 2021. PMID: 34200500 Free PMC article. Review.
References
-
- AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. - PubMed
-
- Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, and T. Horsnell. 1990. Analysis of the coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

