Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration
- PMID: 20573900
- PMCID: PMC2905051
- DOI: 10.1523/JNEUROSCI.0032-10.2010
Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration
Abstract
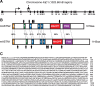
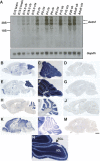
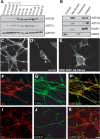
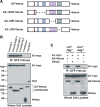
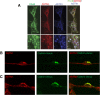
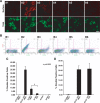
Glial-guided neuronal migration is a key step in the development of laminar architecture of cortical regions of the mammalian brain. We previously reported that neuronal protein astrotactin (ASTN1) functions as a neuron-glial ligand during CNS glial-guided migration. Here, we identify a new Astn family member, Astn2, that is expressed at high levels in migrating, cerebellar granule neurons, along with Astn1, at developmental stages when glial-guided migration is ongoing. Biochemical and flow cytometry experiments show that ASTN2 forms a complex with ASTN1 and regulates surface expression of ASTN1. Live imaging of Venus-tagged ASTN1 in migrating cerebellar granule cells reveals the intracellular trafficking of ASTN1-Venus, with ASTN1-Venus accumulating in the forward aspect of the leading process where new sites of adhesion will form. Treatment of migrating neurons with Dynasore, a soluble noncompetitive inhibitor of Dynamin, rapidly arrests the migration of immature granule cells in a reversible manner, suggesting the critical importance of receptor trafficking to neuronal locomotion along Bergmann glial fibers in the developing cerebellum. Together, these findings suggest that ASTN2 regulates the levels of ASTN1 in the plasma membrane and that the release of neuronal adhesions to the glial fiber during neuronal locomotion involves the intracellular trafficking of ASTN1.
Figures



References
-
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. - PubMed
-
- Anton ES, Marchionni MA, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124:3501–3510. - PubMed
-
- Barkovich AJ, Kuzniecky RI, Dobyns WB, Jackson GD, Becker LE, Evrard P. Malformations of the cortical development. Neuropediatrics. 1996;27:59–63. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
