Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome
- PMID: 20574068
- PMCID: PMC2943146
- DOI: 10.1126/scitranslmed.3000813
Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome
Abstract
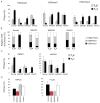
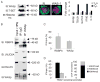
The clinical symptomatology in the X-linked Wiskott-Aldrich syndrome (WAS), a combined immunodeficiency and autoimmune disease resulting from WAS protein (WASp) deficiency, reflects the underlying coexistence of an impaired T helper 1 (TH1) immunity alongside intact TH2 immunity. This suggests a role for WASp in patterning T(H) subtype immunity, yet the molecular basis for the TH1-TH2 imbalance in human WAS is unknown. We have discovered a nuclear role for WASp in the transcriptional regulation of the TH1 regulator gene TBX21 at the chromatin level. In primary TH1-differentiating cells, a fraction of WASp is found in the nucleus, where it is recruited to the proximal promoter locus of the TBX21 gene, but not to the core promoter of GATA3 (a TH2 regulator gene) or RORc (a TH17 regulator gene). Genome-wide mapping demonstrates association of WASp in vivo with the gene-regulatory network that orchestrates TH1 cell fate choice in the human TH cell genome. Functionally, nuclear WASp associates with H3K4 trimethyltransferase [RBBP5 (retinoblastoma-binding protein 5)] and H3K9/H3K36 tridemethylase [JMJD2A (Jumonji domain-containing protein 2A)] proteins, and their enzymatic activity in vitro and in vivo is required for achieving transcription-permissive chromatin dynamics at the TBX21 proximal promoter in primary differentiating TH1 cells. During TH1 differentiation, the loss of WASp accompanies decreased enrichment of RBBP5 and, in a subset of WAS patients, also of filamentous actin at the TBX21 proximal promoter locus. Accordingly, human WASp-deficient TH cells, from natural mutation or RNA interference-mediated depletion, demonstrate repressed TBX21 promoter dynamics when driven under TH1-differentiating conditions. These chromatin derangements accompany deficient T-BET messenger RNA and protein expression and impaired TH1 function, defects that are ameliorated by reintroducing WASp. Our findings reveal a previously unappreciated role of WASp in the epigenetic control of T-BET transcription and provide a new mechanism for the pathogenesis of WAS by linking aberrant histone methylation at the TBX21 promoter to dysregulated adaptive immunity.
Conflict of interest statement
Figures





References
-
- Derry JM, Ochs HD, Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;78:635–644. - PubMed
-
- Imai K, Morio T, Zhu Y, Jin Y, Itoh S, Kajiwara M, Yata J, Mizutani S, Ochs HD, Nonoyama S. Clinical course of patients with WASP gene mutations. Blood. 2004;103:456–464. - PubMed
-
- Trifari S, Sitia G, Aiuti A, Scaramuzza S, Marangoni F, Guidotti LG, Martino S, Saracco P, Notarangelo LD, Roncarolo MG, Dupré L. Defective Th1 cytokine gene transcription in CD4+ and CD8+ T cells from Wiskott-Aldrich syndrome patients. J Immunol. 2006;177:7451–7461. - PubMed
-
- Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott–Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

