MicroRNAs: small RNAs with big effects
- PMID: 20574417
- PMCID: PMC3094098
- DOI: 10.1097/TP.0b013e3181e913c2
MicroRNAs: small RNAs with big effects
Abstract
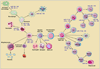
MicroRNAs (miRNAs) are evolutionarily conserved, small ( approximately 20-25 nucleotides), single-stranded molecules that suppress the expression of protein-coding genes by translational repression, messenger RNA degradation, or both. More than 700 miRNAs have been identified in the human genome. Amazingly, a single miRNA can regulate the expression of hundreds of mRNAs or proteins within a cell. The small RNAs are fast emerging as master regulators of innate and adaptive immunity and likely to play a pivotal role in transplantation. The clinical application of RNA sequencing ("next-generation sequencing") should facilitate transcriptome profiling at an unprecedented resolution. We provide an overview of miRNA biology and their hypothesized roles in transplantation.
Conflict of interest statement
Figures

References
-
- Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451(7177):414. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843. - PubMed
-
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

