Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers
- PMID: 20574448
- PMCID: PMC3262678
- DOI: 10.1038/nrc2876
Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers
Abstract
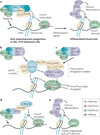
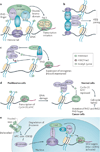
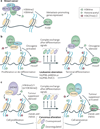
Post-translational modification of histones provides an important regulatory platform for processes such as gene transcription and DNA damage repair. It has become increasingly apparent that the misregulation of histone modification, which is caused by the deregulation of factors that mediate the modification installation, removal and/or interpretation, actively contributes to human cancer. In this Review, we summarize recent advances in understanding the interpretation of certain histone methylations by plant homeodomain finger-containing proteins, and how misreading, miswriting and mis-erasing of histone methylation marks can be associated with oncogenesis and progression. These observations provide us with a greater mechanistic understanding of epigenetic alterations in human cancers and might also help direct new therapeutic interventions in the future.
Figures
References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med. 2007;13:363–372. - PubMed
-
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

