Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells
- PMID: 20574810
- PMCID: PMC2943068
- DOI: 10.1007/s00109-010-0642-1
Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells
Abstract
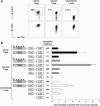
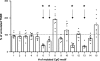
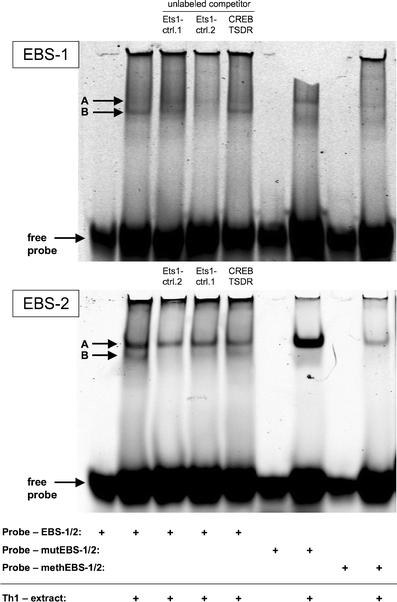
The forkhead-box protein P3 (Foxp3) is a key transcription factor for the development and suppressive activity of regulatory T cells (Tregs), a T cell subset critically involved in the maintenance of self-tolerance and prevention of over-shooting immune responses. However, the transcriptional regulation of Foxp3 expression remains incompletely understood. We have previously shown that epigenetic modifications in the CpG-rich Treg-specific demethylated region (TSDR) in the Foxp3 locus are associated with stable Foxp3 expression. We now demonstrate that the methylation state of the CpG motifs within the TSDR controls its transcriptional activity rather than a Treg-specific transcription factor network. By systematically mutating every CpG motif within the TSDR, we could identify four CpG motifs, which are critically determining the transcriptional activity of the TSDR and which serve as binding sites for essential transcription factors, such as CREB/ATF and NF-κB, which have previously been shown to bind to this element. The transcription factor Ets-1 was here identified as an additional molecular player that specifically binds to the TSDR in a demethylation-dependent manner in vitro. Disruption of the Ets-1 binding sites within the TSDR drastically reduced its transcriptional enhancer activity. In addition, we found Ets-1 bound to the demethylated TSDR in ex vivo isolated Tregs, but not to the methylated TSDR in conventional CD4(+) T cells. We therefore propose that Ets-1 is part of a larger protein complex, which binds to the TSDR only in its demethylated state, thereby restricting stable Foxp3 expression to the Treg lineage.
Figures





Similar articles
-
Mbd2 promotes foxp3 demethylation and T-regulatory-cell function.Mol Cell Biol. 2013 Oct;33(20):4106-15. doi: 10.1128/MCB.00144-13. Epub 2013 Aug 26. Mol Cell Biol. 2013. PMID: 23979593 Free PMC article.
-
The Treg-specific demethylated region stabilizes Foxp3 expression independently of NF-κB signaling.PLoS One. 2014 Feb 5;9(2):e88318. doi: 10.1371/journal.pone.0088318. eCollection 2014. PLoS One. 2014. PMID: 24505473 Free PMC article.
-
Hypomethylation of the Treg-Specific Demethylated Region in FOXP3 Is a Hallmark of the Regulatory T-cell Subtype in Adult T-cell Leukemia.Cancer Immunol Res. 2016 Feb;4(2):136-45. doi: 10.1158/2326-6066.CIR-15-0148. Epub 2015 Dec 17. Cancer Immunol Res. 2016. PMID: 26681759
-
Epigenetic mechanisms of regulation of Foxp3 expression.Blood. 2009 Oct 29;114(18):3727-35. doi: 10.1182/blood-2009-05-219584. Epub 2009 Jul 29. Blood. 2009. PMID: 19641188 Free PMC article. Review.
-
Transcriptional and epigenetic basis of Treg cell development and function: its genetic anomalies or variations in autoimmune diseases.Cell Res. 2020 Jun;30(6):465-474. doi: 10.1038/s41422-020-0324-7. Epub 2020 May 4. Cell Res. 2020. PMID: 32367041 Free PMC article. Review.
Cited by
-
Regulatory T cell function in autoimmune disease.J Transl Autoimmun. 2021 Oct 30;4:100130. doi: 10.1016/j.jtauto.2021.100130. eCollection 2021. J Transl Autoimmun. 2021. PMID: 35005594 Free PMC article. Review.
-
T Helper Cell Lineage-Defining Transcription Factors: Potent Targets for Specific GVHD Therapy?Front Immunol. 2022 Jan 5;12:806529. doi: 10.3389/fimmu.2021.806529. eCollection 2021. Front Immunol. 2022. PMID: 35069590 Free PMC article. Review.
-
Control of Foxp3 induction and maintenance by sequential histone acetylation and DNA demethylation.Cell Rep. 2021 Dec 14;37(11):110124. doi: 10.1016/j.celrep.2021.110124. Cell Rep. 2021. PMID: 34910919 Free PMC article.
-
Treg functional stability and its responsiveness to the microenvironment.Immunol Rev. 2014 May;259(1):115-39. doi: 10.1111/imr.12172. Immunol Rev. 2014. PMID: 24712463 Free PMC article. Review.
-
MeCP2 enforces Foxp3 expression to promote regulatory T cells' resilience to inflammation.Proc Natl Acad Sci U S A. 2014 Jul 8;111(27):E2807-16. doi: 10.1073/pnas.1401505111. Epub 2014 Jun 23. Proc Natl Acad Sci U S A. 2014. PMID: 24958888 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

