Conformational transitions in human AP endonuclease 1 and its active site mutant during abasic site repair
- PMID: 20575528
- PMCID: PMC4676079
- DOI: 10.1021/bi100769k
Conformational transitions in human AP endonuclease 1 and its active site mutant during abasic site repair
Abstract
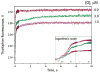
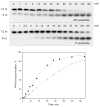
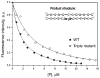
AP endonuclease 1 (APE1) is a crucial enzyme of the base excision repair pathway (BER) in human cells. APE1 recognizes apurinic/apyrimidinic (AP) sites and makes a nick in the phosphodiester backbone 5' to them. The conformational dynamics and presteady-state kinetics of wild-type APE1 and its active site mutant, Y171F-P173L-N174K, have been studied. To observe conformational transitions occurring in the APE1 molecule during the catalytic cycle, we detected intrinsic tryptophan fluorescence of the enzyme under single turnover conditions. DNA duplexes containing a natural AP site, its tetrahydrofuran analogue, or a 2'-deoxyguanosine residue in the same position were used as specific substrates or ligands. The stopped-flow experiments have revealed high flexibility of the APE1 molecule and the complexity of the catalytic process. The fluorescent traces indicate that wild-type APE1 undergoes at least four conformational transitions during the processing of abasic sites in DNA. In contrast, nonspecific interactions of APE1 with undamaged DNA can be described by a two-step kinetic scheme. Rate and equilibrium constants were extracted from the stopped-flow and fluorescence titration data for all substrates, ligands, and products. A replacement of three residues at the enzymatic active site including the replacement of tyrosine 171 with phenylalanine in the enzyme active site resulted in a 2 x 10(4)-fold decrease in the reaction rate and reduced binding affinity. Our data indicate the important role of conformational changes in APE1 for substrate recognition and catalysis.
Figures
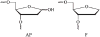




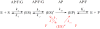
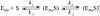
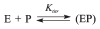
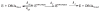
Similar articles
-
Conformational dynamics of abasic DNA upon interactions with AP endonuclease 1 revealed by stopped-flow fluorescence analysis.Biochemistry. 2012 Feb 14;51(6):1306-21. doi: 10.1021/bi201444m. Epub 2012 Jan 30. Biochemistry. 2012. PMID: 22243137
-
The role of Asn-212 in the catalytic mechanism of human endonuclease APE1: stopped-flow kinetic study of incision activity on a natural AP site and a tetrahydrofuran analogue.DNA Repair (Amst). 2014 Sep;21:43-54. doi: 10.1016/j.dnarep.2014.06.008. DNA Repair (Amst). 2014. PMID: 25038572
-
The role of active-site amino acid residues in the cleavage of DNA and RNA substrates by human apurinic/apyrimidinic endonuclease APE1.Biochim Biophys Acta Gen Subj. 2020 Dec;1864(12):129718. doi: 10.1016/j.bbagen.2020.129718. Epub 2020 Aug 25. Biochim Biophys Acta Gen Subj. 2020. PMID: 32858086
-
Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means.Mutat Res. 2000 Aug 30;460(3-4):211-29. doi: 10.1016/s0921-8777(00)00028-8. Mutat Res. 2000. PMID: 10946230 Review.
-
APE1: A skilled nucleic acid surgeon.DNA Repair (Amst). 2018 Nov;71:93-100. doi: 10.1016/j.dnarep.2018.08.012. Epub 2018 Aug 23. DNA Repair (Amst). 2018. PMID: 30170830 Free PMC article. Review.
Cited by
-
APE1/Ref-1 as an emerging therapeutic target for various human diseases: phytochemical modulation of its functions.Exp Mol Med. 2014 Jul 18;46(7):e106. doi: 10.1038/emm.2014.42. Exp Mol Med. 2014. PMID: 25033834 Free PMC article. Review.
-
3CAPS - a structural AP-site analogue as a tool to investigate DNA base excision repair.Nucleic Acids Res. 2016 Mar 18;44(5):2187-98. doi: 10.1093/nar/gkv1520. Epub 2016 Jan 4. Nucleic Acids Res. 2016. PMID: 26733580 Free PMC article.
-
Common Kinetic Mechanism of Abasic Site Recognition by Structurally Different Apurinic/Apyrimidinic Endonucleases.Int J Mol Sci. 2021 Aug 18;22(16):8874. doi: 10.3390/ijms22168874. Int J Mol Sci. 2021. PMID: 34445579 Free PMC article.
-
Conformational Dynamics of Human ALKBH2 Dioxygenase in the Course of DNA Repair as Revealed by Stopped-Flow Fluorescence Spectroscopy.Molecules. 2022 Aug 4;27(15):4960. doi: 10.3390/molecules27154960. Molecules. 2022. PMID: 35956910 Free PMC article.
-
Activity of Human Apurinic/Apyrimidinic Endonuclease APE1 Toward Damaged DNA and Native RNA With Non-canonical Structures.Front Cell Dev Biol. 2020 Oct 30;8:590848. doi: 10.3389/fcell.2020.590848. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33195255 Free PMC article.
References
-
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. - PubMed
-
- Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. - PubMed
-
- Lindahl T. Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat Res. 2000;462:129–135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

